Figure 1.
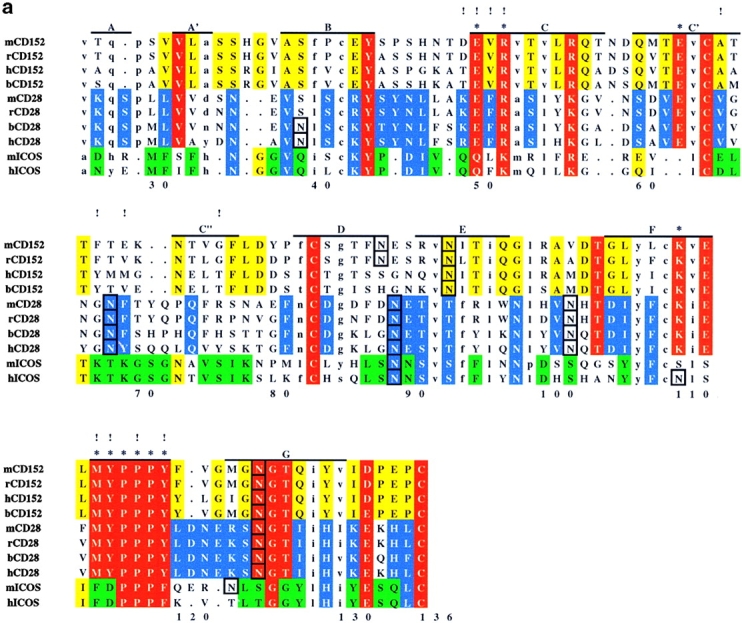
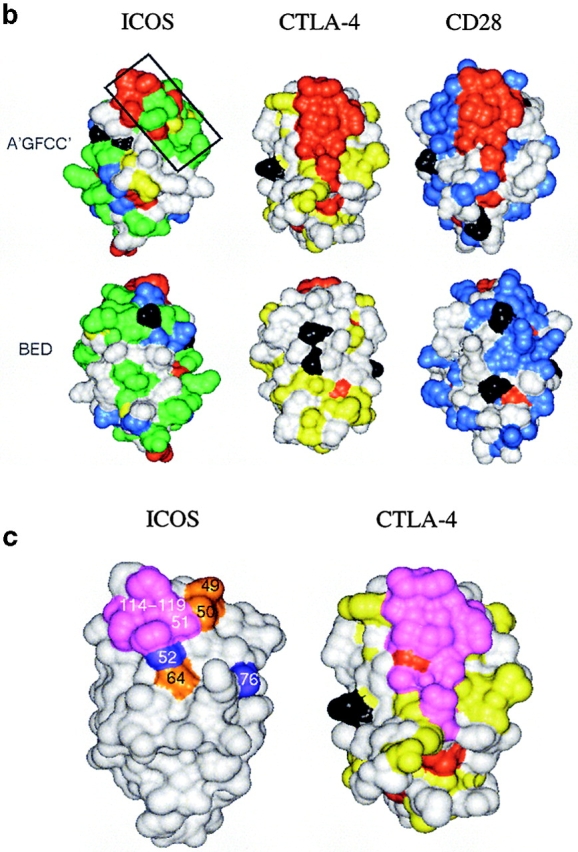
Analysis of residue conservation in the extended CD28 family and mapping of mutagenesis sites. (a) Structure-oriented sequence alignment. Sequences are shown of the extracellular Ig-domains of CD28, CTLA-4, and ICOS from different species. b, bovine; h, human; m, mouse; r, rat. β–strands observed in the solution structure of human CTLA-4 are labeled. Assignments of residues to the A- and C′′-strands are tentative. Residue numbers are given for human ICOS. Ig V-set consensus residues and other hydrophobic core residues are shown in lower case. These residues are important for maintaining structural integrity but are not available for ligand binding. Other residues conserved in CD28, CTLA-4, and ICOS are shown in red. Residues conserved only in CD28 or CD28 and ICOS are shown in blue, residues conserved only CTLA-4 or CTLA-4 and ICOS are shown in yellow, and residues conserved only in ICOS are shown in green. The most conservative residue replacements (e.g., Y/F, R/K, E/Q) are taken into account. Residues conserved in CD28 and CTLA-4 and critical for CD80/CD86 binding are labeled with asterisks. Potential N-linked glycosylation sites are boxed. The positions of ICOS residues subjected to site-specific mutagenesis are labeled with exclamation marks. (b) Three-dimensional analysis of residue conservation. A molecular model of the extracellular domain of human ICOS, the structure of human CTLA-4, and a model of human CD28 are shown in equivalent orientation and with solvent-accessible surface representation. Residues conserved are mapped and color-coded according to a. N-linked glycosylation sites are also mapped and colored black. Two views are shown that focus on the opposite β–sheet surfaces of the Ig domains (top: A′GFCC′ face; bottom: BED face; related by ∼180 degree rotation around the vertical axis). In this orientation, the “PPP” (ICOS) and “MYPPPY” (CD28/CTLA-4) motifs are at the top of the domains and the COOH termini at the bottom. The boxed region in ICOS (top left image) was selected as the likely B7-H2 binding site on the basis of residue conservation. Some ICOS-specific residue conservation is also observed on the BED face of the domain (bottom left). However, this region is masked by a glycosylation site (black), similar to the corresponding regions in CTLA-4 and CD28. (c) Residues important for ligand binding. ICOS and CTLA-4 are shown in the same orientation as in b, and residues are highlighted according to their importance for ligand binding. As identified in previous mutagenesis studies (see reference 26 and references therein), residues on the A′GFCC′ face in CTLA-4 critical for CD80/CD86 binding are colored magenta. All of these residues are strictly conserved in CD28. The positions of ICOS mutants that caused either abolished, reduced, or improved ligand binding are colored in magenta, gold, and blue, respectively. Mutated ICOS residues are numbered.
