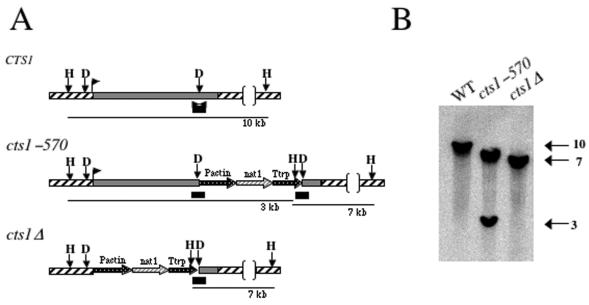
FIG. 2.
Disruption of the CTS1 gene by homologous recombination. (A) The CTS1 gene was disrupted by the insertion of the dominant selectable marker nat1 (nourseothricin resistance) between two NdeI sites in the CTS1 gene, generating the cts1Δ::nat allele, while the cts1-570 truncation mutation was generated by introducing the nat cassette into the 3′ NdeI site corresponding to amino acid 570 of Cts1. The ORF of CTS1 is a solid box, the 5′ and 3′ untranslated regions are indicated by diagonal lines, and the transcription start site is marked by a bent arrow. The 300-bp bar denotes the region used as a probe in panel B. Restriction sites are HindIII (H) and NdeI (D). (B) Southern blot analysis of HindIII-digested genomic DNA from wild-type (WT; JEC21), cts1-570 (DSF22), and cts1Δ (DSF45) strains. Sizes in kilobases are indicated at right.