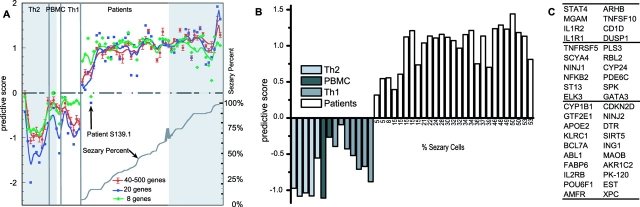
Figure 4.
Classification of low tumor burden CTCL patients using PDA. (A) Effect of decreasing the number of class predictors on classification. Seven measurements are included in the 40–500 genes tracing, 500, 400, 300, 200, 100, 80, and 40 genes. Each point represents the average predictive score for the aforementioned seven classifiers, and the error bars show one standard deviation. Data are shown separately for 20- and 8-gene classifiers. The lower curve shows the percent Sezary cells in each analyzed sample. For each classifier, the predictive scores are normalized by the average predictive score of patients. The shaded areas indicate values for replicate samples from the training set. (B) Classification of 27 low CTCL patients and 14 controls with 40 genes (PBMCs = untreated). Negative values indicate the expression profiles that define the negative or control class and positive values identify samples assigned to the positive or patient class. The height of the columns measure how well each sample is classified by the list of 40 genes. Percent Sezary cells are indicated for each sample. (C) List of the 40 best classifiers. Genes are listed top to bottom, sorted by the value of their predictive power. Genes up-regulated in patients are in the right column; genes down-regulated in patients are in the left column.