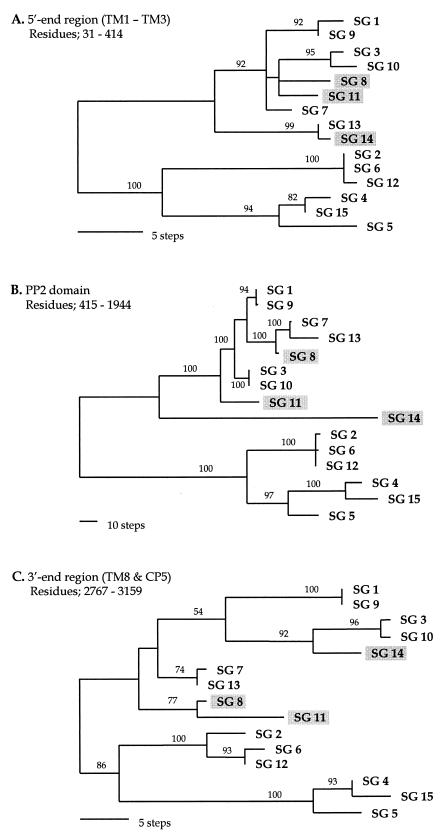
FIG. 4.
Gene phylogenies inferred from three regions of the dotA gene. These trees were constructed from nucleotide sequences by parsimony analysis. (A) One of the six most parsimonious trees (87 steps) inferred from the 5′-end regions corresponding to TM1 to TM3 (residues 31 to 414); CI = 0.851; RI = 0.930. (B) The unique parsimonious tree (531 steps) constructed from the PP2 domain of residues 415 to 1944; CI = 0.887; RI = 0.948. (C) One of the four most parsimonious trees from the 3′-end regions, corresponding to the TM8 and CP5 domains (residues 2767 to 3159); CI = 0.748; RI = 0.845. The branch lengths are proportional to changes in the nucleotides. The numbers on the branches are the percentages of support from bootstrap analysis (500 replications). The three strains that have different positions in the three phylogenies are shaded.