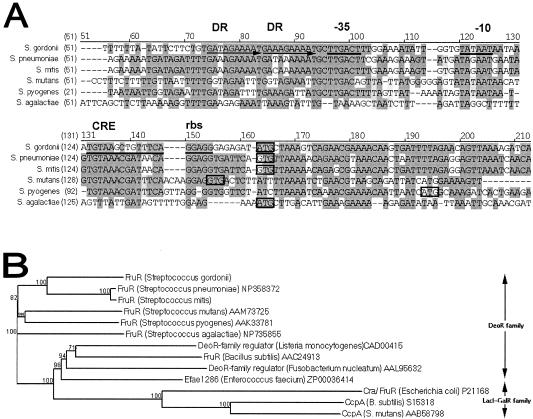
FIG. 2.
(A) Alignment of the promoter region of S. gordonii fruR with the fruR homologs in S. pneumoniae (accession no. NP345362), S. mitis, S. mutans (accession no. NC004350), S. pyogenes (accession no. 002737), and S. agalactiae (accession no. NC004116) determined by using the AlignX program in the Vector NTI software (Informax). Streptococcal sequences without accession numbers were obtained from the unfinished microbial genome database (www.ncbi.nlm.nih.gov). Nucleotides that are identical and conserved are indicated by dark grey and light grey backgrounds, respectively, and start codons are enclosed in boxes. In S. gordonii, direct repeat regions (DR) are indicated by horizontal arrows, while the CRE, putative ribosome binding site (rbs), putative −35, and putative −10 regions are underlined. (B) Phylogenetic tree of S. gordonii FruR and other bacterial sugar PTS regulatory proteins. The phylogenetic relatedness dendrogram was constructed on the basis of amino acid sequence similarities by using the AlignX program in the Vector NTI software (Informax Inc.), which utilizes the neighbor-joining algorithm (32). The reliability of the topology was estimated by performing 100 bootstrap trials, and the bootstrap values are expressed in percentages at branch points (www.genebee.msu.su). Accession numbers follow the species names, and sequences without accession numbers were obtained from the unfinished microbial genome sequence database (www.ncbi.nlm.nih.gov).