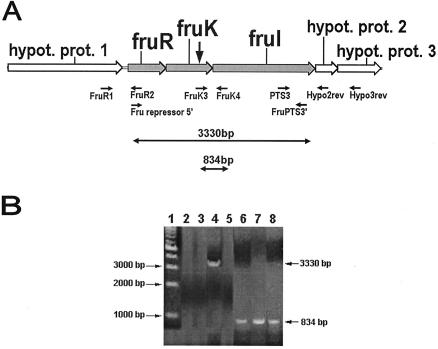
FIG. 3.
RT-PCR analysis of RNA extracted from the S. gordonii Challis 2, fruK::Tn917-lac, and fruK/fruR strains. (A) Organization of the fructose PTS operon and adjacent genes, locations of primers, and predicted size of the successfully amplified RT-PCR product. See Table 1 for the primers used. The vertical arrow indicates the position of the Tn917-lac insertion in the biofilm-defective mutant. hypot. prot., hypothetical protein. (B) RT-PCR products obtained by using total RNA extracted from the Challis 2 strain, the fruK::Tn917-lac mutant, or the fruK/fruR double mutant as the template. Lane 1, 1-kb DNA marker; lane 2, Challis 2 RNA with primers FruR1 and FruR2; lane 3, Challis 2 RNA with primers PTS3 and Hypo2rev; lane 4, Challis 2 RNA with primers Fru repressor5′ and FruPTS3′; lane 5, Challis 2 RNA with primers PTS3 and Hypo3rev; lane 6, Challis 2 RNA with primers FruK3 and FruK4; lane 7, fruK::Tn917-lac RNA with primers FruK3 and FruK4; lane 8, fruK/fruR RNA with primers FruK3 and FruK4.