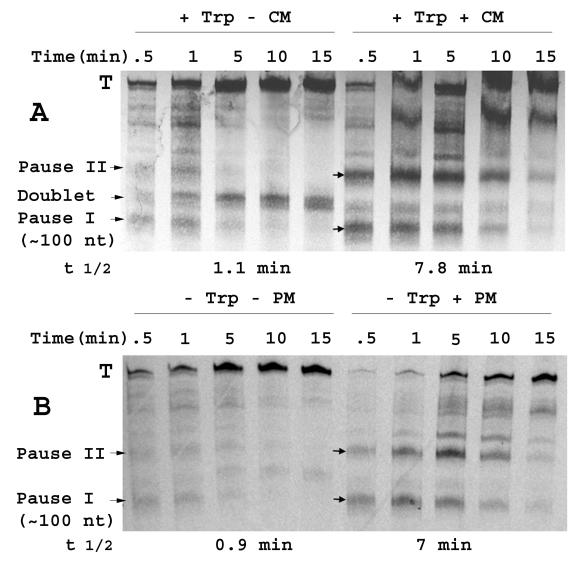
FIG. 2.
(A) Single-round transcription analyses examining the effects of chloramphenicol on transcription pausing. The DNA template used to direct the S-30 system was a self-ligated PCR fragment containing the region from positions −195 to + 115 of the tna operon (relative to the tna transcription initiation site), followed by the rpoBC transcription terminator sequence. The corresponding mRNA sequence is shown in Fig. 1A. S-30 reaction mixtures (50-μl volume) with 1 mM tryptophan and 50 μg of bicyclomycin per ml but without CTP, GTP, and UTP were incubated at 37°C for 10 min. Then 20 μCi of [α-33P]UTP and 200 μg of rifampin per ml with (+) or without (−) 200 μg of chloramphenicol (CM) per ml were added together to the reaction mixture. Samples were taken at the indicated time points, the reactions were stopped by phenol extraction, and the samples were loaded on a urea-polyacrylamide RNA gel. Pauses that generate RNA species longer than 115 nt (pause II, etc.) correspond to hairpin structures that could form in the transcript produced from the incorporated rpoBC terminator (Fig. 1A); these pauses are not directly relevant to this study. The positions of the terminated (T) transcripts, RNA doublet, and transcript produced by pausing at the end of the tnaC coding region (pause I), are indicated. The half-life (t 1/2) of pause I was measured using a phosphorimager (7). (B) Effect of puromycin addition on transcription pausing. Single-round assays were conducted as described above for panel A in the absence of a high level of tryptophan. In addition, chloramphenicol was replaced by 200 μg of puromycin (PM) per ml, where indicated. The half-life (t 1/2) of pause I was measured using a phosphorimager (7).