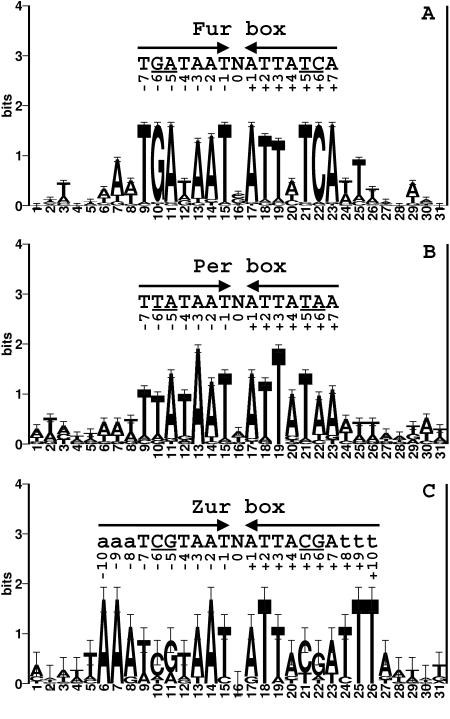
FIG. 1.
(A) DNA sequence logo of the B. subtilis Fur DNA binding sites (3, 44). A set of 40 DNA sequences (20 sites together with their reverse complements) containing Fur box sites were aligned with CLUSTALW (47), and then a window of 31 bases around the 7-1-7 motif was used to generate a sequence logo. The height of each letter corresponds to its relative abundance at that position. The letters above the logo show the sequence of conserved bases. The underlined letters represent bases that are different among Fur, PerR, and Zur boxes. (B) Sequence logo of the B. subtilis Per box. A total of 12 Per boxes were used for alignment. (C) DNA logo of B. subtilis Zur DNA binding sites. Note that there are only four known Zur boxes found in the B. subtilis genome.