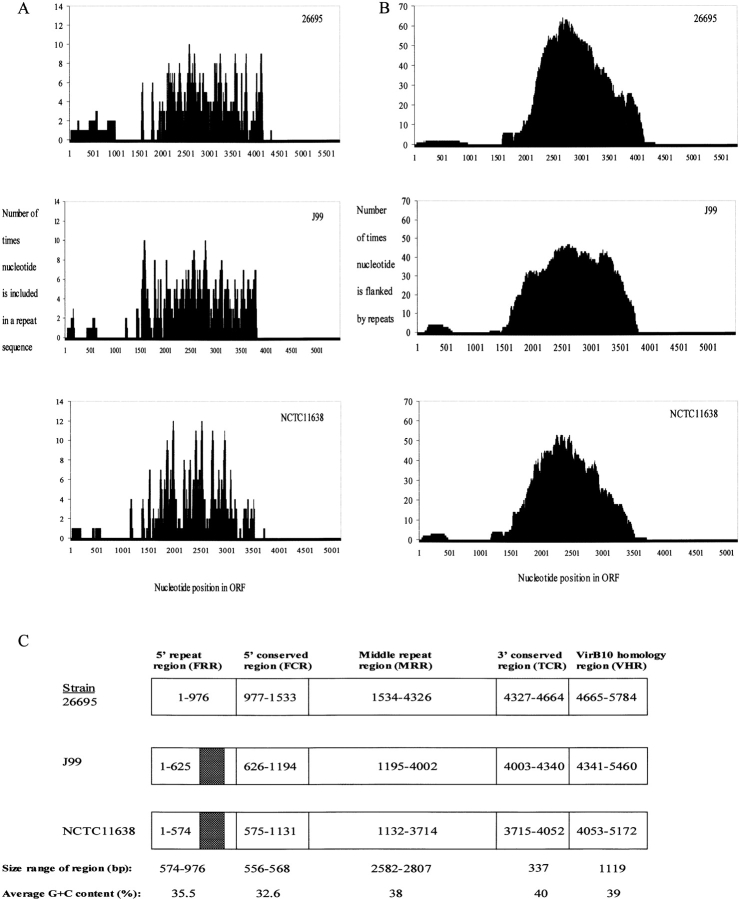
Figure 1.
Direct nucleotide repeats in the structure of H. pylori cagY. (A) The abscissa represents the nucleotide position in each ORF and the ordinate for each of three strains (26695, J99, NCTC11168) represents the number of times each nucleotide is part of a direct repeat sequence ≥16 nucleotides. For each strain, the repeats are clustered in two regions. (B) The abscissa represents the nucleotide position in each ORF and the ordinate represents the number of times that identical repeats ≥16 bp flank each nucleotide. The presence of a nucleotide between identical repeats indicates that it could be deleted or duplicated if recombination occurred between the repeats. (C) For the three strains analyzed, cagY might be separated into five defined regions present in each sequence, which we termed the FRR, the FCR, the MRR, the TCR, and the VHR. None of the repeats are shared in both the FRR and MRR. Downstream of both the FRR and MRR are highly conserved regions of ∼550 bp (FCR and TCR), followed by the VHR portion of cagY, encoding a polypeptide highly homologous to the A. tumefaciens virB10 product. The FCR, TCR, and VHR have high levels of DNA identity between strains, with FCR size differences due to small variations in the boundaries of the adjacent repeat regions. FRR size variation is largely due to a 329-bp deletion (hatched region). The G+C content is lowest and deviates substantially from the remainder of the genome in the portion of cagY upstream of the MRR.