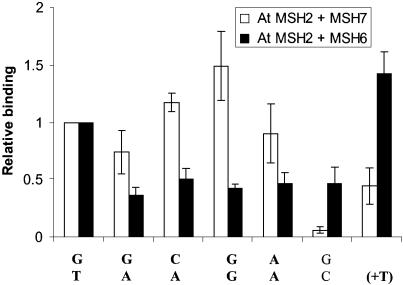
Figure 3.
Mismatch-recognition spectra for AtMutSα and AtMutSγ. Electrophoretic mobility shift analyses of mixtures of 0.1 pmol of indicated 51 bp DNA duplexes with equal aliquots of co-synthesized AtMSH7 + AtMSH2 (AtMutSγ) or AtMSH6 + AtMSH2 (AtMutSα) polypeptide mixtures were described in the Figure 2 legend. Summed intensities of mismatch-specific DNA–protein bands (designated by letter A in Fig. 2) for each mismatch were divided by intensities for G/T DNA. Data correspond to means for three independent determinations and standard deviations for binding to MutSγ (open bars), or MutSα (filled bars). Oligomer (upper/lower) pairs used were as indicated in the Figure 2 legend, plus those for G/G (1/16) and A/A (1/17).