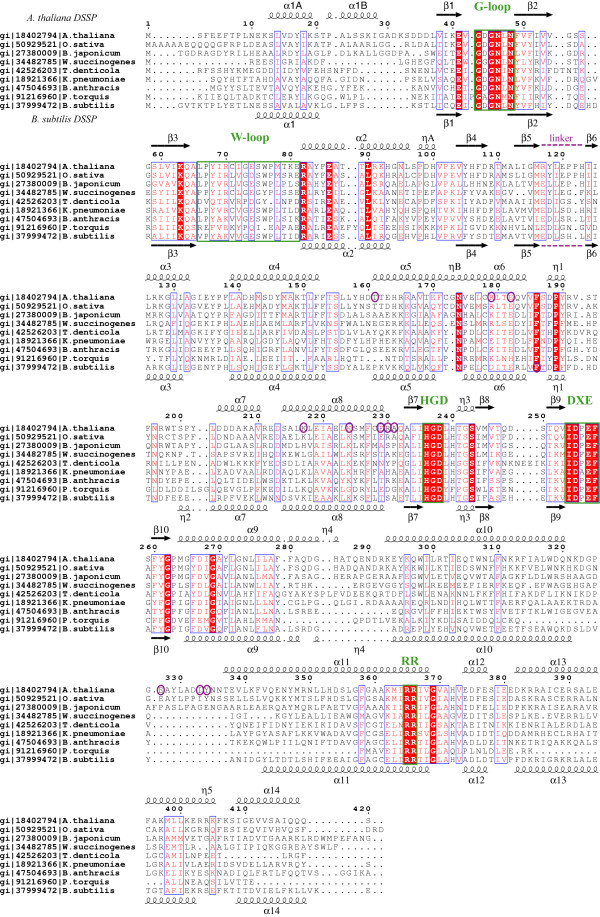
Figure 1.
Multiple sequence alignment of selected MTR kinase sequences mapping the secondary structural elements found in A. thaliana (top) and B. subtilis (bottom) MTR kinase structures. The protein sequences aligned are from Arabidopsis thaliana, Oryza sativa, Bradyrhizobium japonicum, Wolinella succinogenes, Treponema denticola, Klebsiella pneumoniae, Bacillus anthracis, Psychroflexus torquis and Bacillus subtilis. Their gene identifiers (gi) from the National Center for Biotechnology Information (NCBI) are also listed. The secondary structural elements were defined according to DSSP [48], numbered by the order of their appearance, and named to be consistent between the two enzymes. α helices are presented as curly lines and β strands by arrows. η stands for a 310 helix. According to DSSP, the A. thaliana enzyme does not have the η2 helix found in the B. subtilis enzyme. The multiple sequence alignment was performed using the program T-Coffee [49], and the figure prepared using ESPript [50]. Strictly conserved residues are denoted in white and framed in blue boxes with a red background; residues conserved in at least 70% of the sequences are denoted in red and framed in blue boxes with a white background. The G-loop, the W-loop, the Mg-binding DXE-motif, the HGD catalytic loop, and the MTR-binding RR-motif are framed in green boxes. Residues circled in purple are involved in dimer formation. The linker region connecting the N-lobe and C-lobe is also indicated.