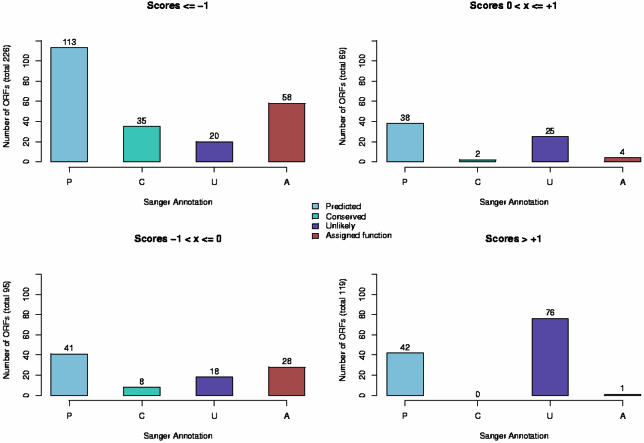
Figure 5.
Breakdown of ORFs based on Sanger Institute annotation versus LDA based annotation. The Sanger Institute provides three sub-classifications of hypothetical annotations. Those ORFs that show sequence-based homology to hypothetical annotations in other organisms are termed ‘conserved hypothetical protein’ (denoted here as ‘Conserved’ or ‘C’). If an ORF is on the opposite strand from the predicted coding strand or has unusual GC composition, it is sometimes labeled ‘hypothetical protein, unlikely’ (based on details provided on the Sanger Institute web pages and EMBL entry). We have shown these here as ‘Unlikely’ or ‘U’. Finally, some ORFs are annotated merely as ‘hypothetical protein’ and we refer to these as ‘predicted’ or ‘P’. For those ORFs with a function assignment, we use the term ‘Assigned function’ or ‘A’. Plots on the left side of this figure show the distribution of annotations for ORFs that our method would label as likely coding, while plots on the right side if this figure show ORFs that our method would identify as unlikely to be true coding regions.