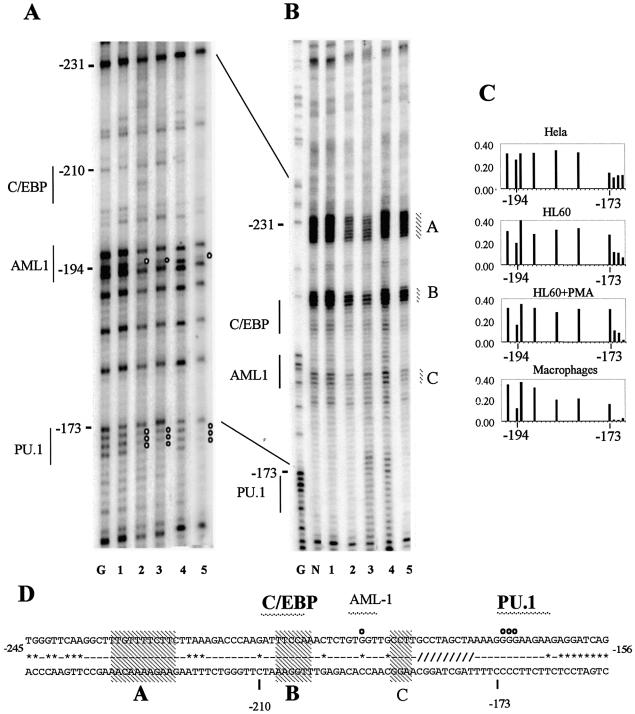
Figure 3.
DMS and UV in vivo footprinting of the human c-FMS promoter reveals transcription factor occupancy with associated alterations in chromatin fine structure in c-FMS expressing cells. (A) In vivo DMS footprinting reveals significant protections over the PU.1 and AML-1 sites previously identified with in vitro assays (20) in c-FMS expressing cells. (B) In vivo UV footprinting reveals significant alterations in DNA topology over three sites (A, B and C) over and upstream from the AML1 and C/EBP sites in c-FMS expressing cells. Both undifferentiated and PMA-differentiated HL-60 cells show similar occupancy and chromatin fine structure to primary macrophages. (C) Example of the quantification of bands generated by in vivo DMS footprinting depicting the region in the c-FMS promoter around the protected guanine at –194 bp at the AML1 site and the region of altered DMS reactivity around the PU.1 site at –173 bp. (D) The promoter sequence with protections indicated as white circles. Sequences conserved between mouse and human are indicated by a line (-); sequence deviations are displayed as *. The region shown as /// indicates a 20 bp poly(A) insert in the mouse sequence. Transcription factor binding sites in bold are conserved between mouse and human. Regions with altered UV pyrimidine dimer formation are indicated (A–C), with conserved dimer changes indicated in bold. Lane labelling for (A) and (B): lane G, G reaction; lane N, naked DNA; lane 1, HeLa; lane 2, HL-60; lane 3, HL-60 + PMA; lane 4, human fibroblasts; lane 5, human macrophages.