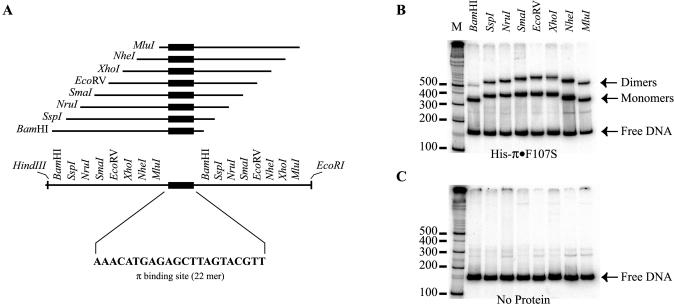
Figure 6.
Iteron DNA binding assay. (A) Construction of a series of DNA fragments with permuted iteron sequence. Blunt end, double-stranded, 22 bp iteron DNA (sequence shown) was generated from synthetic, single-stranded oligos and ligated into HapI-digested pBEND5 (36) giving rise to the plasmid pRK20. Several DNA probes were generated from this plasmid by digestion with restriction enzymes, as indicated, variously placing the iteron sequence near either end or near the middle of the DNA fragment. (B and C) The results of electrophoretic mobility shift assays. The positions of π monomers, dimers and free DNA are indicated by arrows. M is a 32P-labeled 100 bp ladder for DNA fragment migration reference. In (B), DNA fragments were incubated with Rep protein as described in Materials and Methods. (C) is a ‘negative’ control incubated without π protein to reveal any bending that might be intrinsic to the iteron sequence.