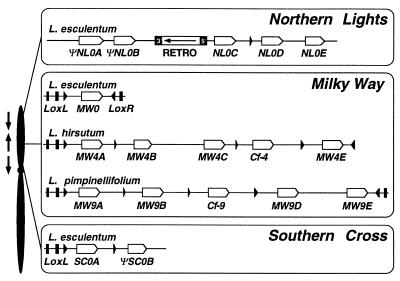
Figure 1.
Map position and physical structure of the NL, MW, and SC clusters of Hcr9 genes. On the left is a schematic genetical map of tomato chromosome 1 showing the position of three Hcr9 loci relative to each other. Arrows indicate the transcriptional polarity of Hcr9s at the different loci—e.g., MW Hcr9s are transcribed toward the telomere which is the opposite direction to Hcr9s at SC and NL. On the right, the physical organization of Hcr9 clusters is shown. All of the displayed haplotypes have been entirely sequenced (5, 7). Open arrows indicate the position and transcriptional polarity of Hcr9s. The haplotypes at MW are all flanked by convergently orientated lipoxygenase genes (LoxL and LoxR). The exons of Lox are indicated by black boxes. The 3′ most exons are shown by black triangles, indicating the transcriptional polarity. Two types of fragments originating from the 3′ end of LoxL are interspersed between Hcr9 genes at MW, SC, and NL, and are also represented by black triangles. Hcr9 pseudogenes are labeled with a Ψ prefix. RETRO denotes a retrotransposon insertion in the NL0 haplotype in which the terminal repeats are shown by black boxes. The transcriptional direction of the polyprotein gene is shown by an arrow.