Abstract
Cellular interleukin 10s (cIL-10s) of human and murine origin have extensive sequence and structural homology to the Epstein-Barr virus BCRF-I gene product, known as viral IL-10 (vIL-10). Although these cytokines share many immunosuppressive properties, vIL-10 lacks several of the immunostimulatory activities of cIL-10 on certain cell types. The molecular and cellular bases for this dichotomy are not currently defined. Here, we show that the single amino acid isoleucine at position 87 of cIL-10 is required for its immunostimulatory function. Substitution of isoleucine in cIL-10 with alanine, which corresponds to the vIL-10 residue, abrogates immunostimulatory activity for thymocytes, mast cells, and alloantigenic responses while preserving immunosuppressive activity for inhibition of interferon γ production and prolongation of cardiac allograft survival. Conversely, substitution of alanine with isoleucine in vIL-10 converts it to a cIL-10–like molecule with immunostimulatory activity. This single conservative residue alteration significantly affects ligand affinity for receptor; however, affinity changes do not necessarily alter specific activities for biologic responses in a predictable fashion. These results suggest complex regulation of IL-10 receptor–ligand interactions and subsequent biological responses. These results demonstrate that vIL-10 may represent a captured and selectively mutated cIL-10 gene that benefits viral pathogenesis by leading to ineffective host immune responses. The ability to manipulate the activity of IL-10 in either a stimulatory or suppressive direction may be of practical value for regulating immune responses for disease therapy, and of theoretical value for determining what aspects of IL-10 activity are important for normal T cell responses.
Keywords: interleukin 10, stimulation, suppression, affinity, signaling
Introduction
IL-10 was originally described as cytokine synthesis inhibitory factor because of its ability to turn off cytokine production by T cells 1. Further investigations demonstrated that the immunosuppressive effects of IL-10 are often at the level of the APC as well as at the level of the T cell 2. Thus, IL-10 inhibits monocyte and macrophage synthesis of IL-1α, IL-1β, IL-6, IL-8, IL-12, TNF-α, GM-CSF, and reactive oxygen and nitrogen intermediates 3 4 5 6. IL-10 can effectively treat the cytokine syndrome and toxicity caused by anti-CD3 mAb or endotoxin by inhibiting the production of proinflammatory cytokines 7 8 9. Autoimmune models of rheumatoid arthritis, thyroiditis, and collagen-induced arthritis and a model of herpetic stromal keratitis all suggest negative regulatory roles for IL-10 in limiting inflammation and immunopathology 10 11 12 13. IL-10–deficient knockout mice have highly polarized Th1 responses and develop a severe colitis related to chronic stimulation by enteric antigens 14. In humans, Crohn's colitis may even be susceptible to treatment with systematically administered IL-10 15. Further reinforcement for the notion that IL-10 is an immunosuppressive cytokine came from the discovery of viral IL-10 (vIL-10) 16, which was shown to have identical immunosuppressive properties to cellular IL-10 (cIL-10), inhibiting IFN-γ production, MHC class II expression, T cell proliferation, and B cell IgE production 17 18 19 20 21 22.
However, cIL-10 also has immunostimulatory properties in some circumstances. It can act as a stimulatory factor for immature and mature thymocytes, mast cells, and B cells 23 24 25. Interestingly, although cIL-10 can costimulate thymocyte and mast cell proliferation and B cell MHC class II expression, vIL-10 cannot 18 22 23, suggesting differences in the structure and function of the two molecules. Transduction of tumors with a retroviral vector encoding mouse (m)IL-10 results in enhanced antitumor immunity and rejection, whereas vIL-10 tumor cell transduction results in immune suppression and tumor growth 26. Our previous studies showed that rejection of mouse heart allografts was substantially inhibited when the grafts were induced by retroviral gene transfer to express vIL-10, but not mIL-10 27. These observations suggest that vIL-10 has preserved only a subset of the activities of cIL-10 and imply the existence of at least two functional domains, stimulatory and suppressive, only the latter of which has been conserved by EBV. The structural basis for the two activities within cIL-10, or between cIL-10 and vIL-10, has not been elucidated. Using structure–function analysis, we now report that the single amino acid isoleucine at position 87 of cIL-10 is critical for immunostimulatory activity. The finding that a single amino acid can determine the range of immunological properties of IL-10 provides a structural lead for defining the separate functions of IL-10 and should facilitate the dissection of the cellular and molecular events that are regulated by the IL-10 molecule. Moreover, it may provide a basis for the design of novel IL-10–related therapeutic reagents for clinical application.
Materials and Methods
Chimeric IL-10 Constructs and Transfection.
mIL-10 and vIL-10 were cloned into the pMP6A expression vector, which has a CMVie promoter (28; Applied Immune Sciences, Inc.). A series of chimeric mIL-10/vIL-10 cDNAs was constructed and cloned by PCR-based site-directed mutagenesis 29. The 5′ external primer containing a HindIII site, and 3′ external primer containing a BamHI site were synthesized and used to amplify the relevant region. The internal primers at the specific sites of mIL-10 or vIL-10 joining were synthesized and phosphorylated, mIL-10 PCR fragments were ligated to vIL-10 PCR fragments, the ligation products were digested with HindIII and BamHI, and the final product was ligated to pMP6A. For point mutations, internal primers containing specific mutations were used for PCR. Some human IL-10 (hIL-10) constructs were cloned into the pcDNA3.1myc-his vector (Invitrogen). All constructs were sequenced to confirm fidelity. Purified chimeric cDNA plasmids were transfected into COS-7 cells by calcium phosphate method, and the COS cell supernatants were harvested after 48 h. Protein expression was analyzed by internal [35S]methionine labeling 16. In brief, cell cultures on the second day after transient transfection were cultured in 3 ml of methionine-deficient medium supplemented with [35S]methionine (Amersham Pharmacia Biotech) at 100 μCi/ml and tunicamycin at 170 ng/ml for 48 h. Supernatants were concentrated 10-fold and analyzed by 14% SDS-PAGE. For Western blotting, COS cell supernatants of COOH-terminal myc-his–tagged hIL-10 were concentrated fivefold, run on an 8–16% SDS-PAGE gel, transferred to a nitrocellulose membrane, blotted with anti-myc mAb (1:10,000; Invitrogen), and developed sequentially with anti–mouse IgG-horseradish peroxidase (1:7,500; Amersham Pharmacia Biotech) and enhanced chemiluminescent reagents (Amersham Pharmacia Biotech).
Cell Culture, Proliferation Assays, and IFN-γ Inhibition.
1 × 105 thymocytes from CBA/J mice were incubated in complete RPMI medium (RPMI 1640 supplemented with 10% FCS, 1 mM sodium pyruvate, 2 mM l-glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, 1× nonessential amino acids, and 2 × 10−5 M 2-ME) with mIL-2 (500 U/ml; PharMingen), mIL-4 (250 U/ml; PharMingen), and various amounts (1–20%) of COS cell supernatants for 3–5 d 22. Proliferation was assessed by 18-h [3H]thymidine incorporation. The MC/9 cell line was purchased from American Type Culture Collection and grown in RPMI plus 10% FCS and 5% mouse ConA supernatants. MC/9 cells were rested in complete media overnight, then 1 × 105 cells per well were incubated with various concentrations of IL-10 COS supernatants for 24 h, and proliferation was assessed by 6-h [3H]thymidine incorporation 24. Ba/F3 cells expressing recombinant mIL-10R1 were a gift from Dr. K. Moore (DNAX, Inc., Palo Alto, CA) and maintained as described 29. Proliferation of Ba/F3-mIL-10R1 transfectants in response to IL-10 was analyzed as described 30 31 with a colorimetric assay using Alamar blue (Accurate Chemical). For inhibition of IFN-γ production, PBMCs were purified from healthy donors using Ficoll-Paque Plus (Amersham Pharmacia Biotech), 2 × 105 cells per well were incubated with soluble OKT3 (0.1 μg/ml) plus various concentrations of IL-10 COS supernatants for 48 h, and IFN-γ production was measured by two-antibody capture ELISA (PharMingen) and compared with an IFN-γ standard (PharMingen). The concentration of various IL-10s that induced half-maximal responses in each of the bioassays was defined as 1 U/ml and used to compare specific activities of the various constructs and the different assays.
Cardiac Transplantation.
Donor neonatal C57BL/6 mice were killed, and whole hearts were removed, injected with 0.31 μg of various IL-10 plasmid DNA constructs complexed with 10 μg of dendrimer G5-EDA to enhance transfection 32, and transplanted into CBA/J recipients. Survival of cardiac allografts was followed with EKG monitoring every other day. Cessation of cardiac electrical activity was the determinant of rejection. P values were calculated by Student's t test.
Induction of HLA-B7 and Electrophoretic Mobility Shift Assay of Signal Transducers and Activators of Transcription.
The 16-9 Chinese hamster ovary (CHO) cells transfected with an HLA-B7 reporter construct and expressing hIL-10R1/hIFN-γR1 chimera, hIL-10R2, or hIL-10R1/hIFN-γR1 plus hIL-10R2 33 were maintained in complete F12 medium containing 450 μg/ml G418. 1 × 105 cells were treated with 10% COS supernatants of hIL-10 or hIL-10(I87A) for 72 h. HLA-B7 antigen was detected by treatment of cells with anti-HLA (w6/32) mAb, followed by treatment with FITC-conjugated goat anti–mouse IgG (PharMingen), and 1 × 104 cells were analyzed by FACScan™ (Becton Dickson). For electrophoretic mobility shift assay (EMSA), 2 × 106 cells were exposed to 4 ml 100% COS cell supernatants containing hIL-10 or hIL-10(I87A) for 15 min. The cells were lysed, and nuclear extracts were obtained for binding to 32P-labeled 22-bp IFN-γ activation sequence (GAS) element in the promoter region of the human IFN regulatory factor 1 (IRF-1) gene 33. 1 μl of anti–signal transducer and activator of transcription 1 (Stat1) or anti-Stat3 antibodies (Santa Cruz Biotechnology) was added to the incubation mixture, incubated for 20 min at 22°C, then 4 μl of the reaction mixture was electrophoresed at 200 V for 4 h at 4°C on a 16 × 16 cm 5% polyacrylamide (19:1 acrylamide/bisacrylamide) gel. The dried gel was exposed to Kodak XAR-5 film with an intensifying screen for 2 d at −80°C. When IL-3–dependent derivatives of the Ba/F3-mIL-10R1 cell line were used, 5 × 106 cells were plated in the absence of IL-3, cultured for 4 h, and then stimulated with 4 ml 100% COS cell supernatants containing hIL-10 or hIL-10(I87A) for 15 min. Nuclear extracts and EMSAs were performed as described previously by using the 32P-labeled 18-bp probe based on the IFN-γ response region (GRR) within the promoter of the FcγRI gene 34.
Protein Purification.
hIL-10, hIL-10(I87A), vIL-10, and vIL-10(A87I) were purified by the pMAL Protein Fusion and Purification System (New England Biolabs). PCR fragments were created encoding the mature proteins with EcoRI and BamHI sites at the 5′ and 3′ ends, respectively. The EcoRI-BamHI restriction fragments encoding hIL-10 and hIL-10(I87A) were cloned into pMAL-c2 vector, which leads to cytoplasmic expression of the maltose binding protein (MBP) fusion proteins in Escherichia coli, and the EcoRI-BamHI restriction fragments encoding vIL-10 and vIL-10(A87I) were cloned into pMAL-p2 vector, which leads to periplasmic expression of similar constructs. Fusion proteins were isolated from bacterial cell lysates and purified by maltose affinity column, cleaved by factor Xa, and repurified by Q-Sepharose ion exchange chromatography according to the manufacturer's recommendations. Purified proteins were run on an 8–16% SDS-PAGE gel, analyzed by silver staining, and purity >95% was achieved in all cases. Purified protein concentrations were determined by measuring absorbance at 280 nm by comparison with a calibration curve prepared from measurements with standard hIL-10 (PeproTech) solutions. Endotoxin was <0.1 ng/μg as determined by Endotoxin kit (Sigma Chemical Co.).
125I-labeled hIL-10 Competitive Displacement.
125I-labeled hIL-10 (75–140 μCi/μg) was purchased from NEN Life Science Products. 125I-labeled hIL-10 binding and competitive displacement were assessed as described 31. In brief, 5 × 105 Ba/F3-mIL-10R1 cells were incubated in 200 μl as duplicate samples in 100 pM 125I-labeled hIL-10 in the presence of between 0.6 pM and 200 nM hIL-10, hIL-10(I87A), vIL-10, and vIL-10(A87I) for 4 h at 4°C in RPMI 1640, 2% BSA, 0.02% sodium azide (binding buffer) with shaking. Reaction mixtures were overlaid onto 150 μl of dibutyl phthalate/dioctyl phthalate (3:2) in conical bottom tubes and centrifuged for 1 min (Eppendorf 5413). The pellets were cut off with a razor blade and analyzed in a COBRA® II gamma counter (Packard Instruments).
Results
Isoleucine 87 of cIL-10 Is Critical for Proliferative Activity.
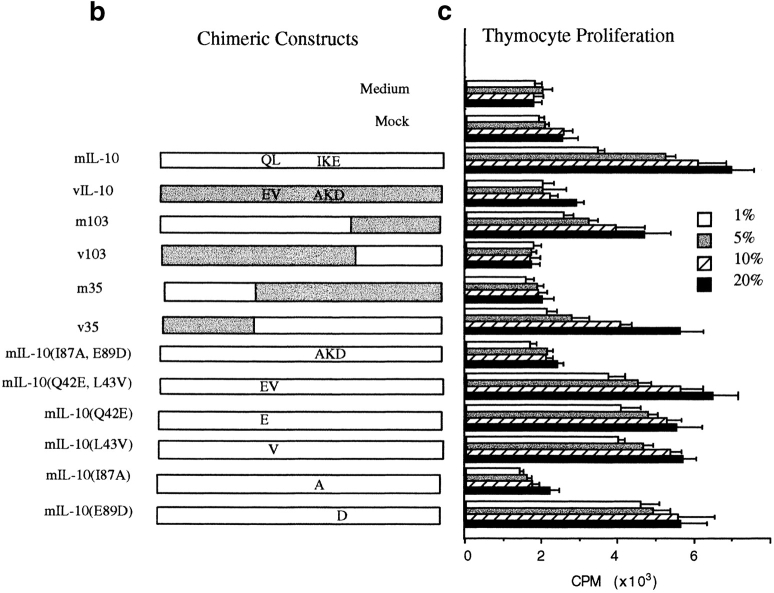
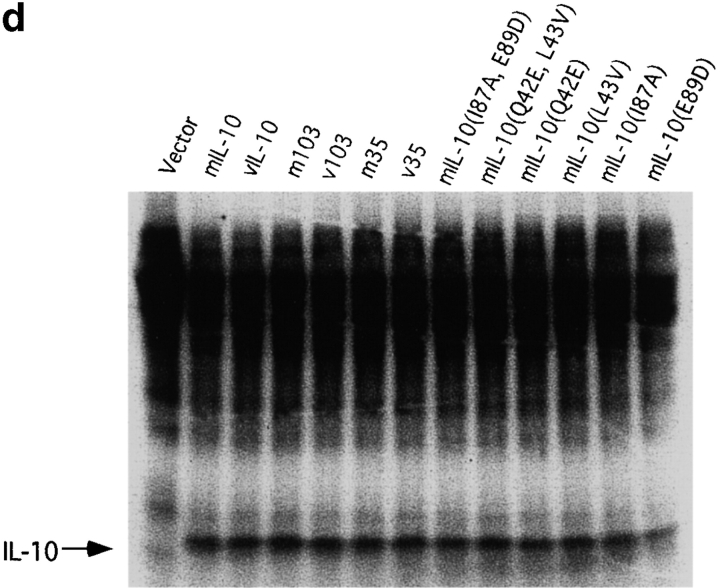
hIL-10 has 84% amino acid identity to viral IL-10 and 73% identity to mIL-10. The linear structure of IL-10 (Fig. 1 a) and crystallographic data show that the greatest differences among vIL-10 and cIL-10s lie in the NH2 terminus, implying that these differences in structure may ultimately lead to changes in immunological function 35 36 37 38. To map which region(s) of the IL-10 molecule determines the immunostimulatory and immunosuppressive activities of the cytokine, we constructed a series of chimeric hybrids that reciprocally exchanged the NH2 or COOH one third of the molecules between mIL-10 and vIL-10 (Fig. 1 b). Because of close molecular homology, it was likely that such exchanges would not destroy overall structure and biological activity, but selectively choose for restricted biological function. Plasmid expression vectors containing these fusion constructs were transfected into COS-7 cells, and the cell supernatants were harvested after 48 h. The expression of all fusion proteins was comparable (Fig. 1 d), as assayed by [35S]methionine labeling. Since mIL-10 has N-linked oligosaccharides, whereas vIL-10 does not, the [35S]methionine labeling was performed in the presence of tunicamycin to eliminate the glycosylation and size differences between the various chimeric constructs. The COS cell supernatants were then tested in the thymocyte coproliferation assay. This is a discordant response, since cIL-10 causes proliferation and vIL-10 does not. The results in Fig. 1 c unexpectedly showed that the difference in activity between mIL-10 and vIL-10 does not reside in the NH2-terminal region of the molecule. Thus, the COOH-terminal 57 amino acids, or NH2-terminal 35 amino acids of mIL-10 could be replaced with the corresponding vIL-10 residues without affecting the costimulation of thymocyte proliferation. Analysis of the proliferative data in Fig. 1 c suggested that the positions responsible for the immunological differences between mIL-10 and vIL-10 reside in the central region of the peptides. Importantly, substitution of large portions of mIL-10 with vIL-10 did not destroy biological activity.
Figure 1.
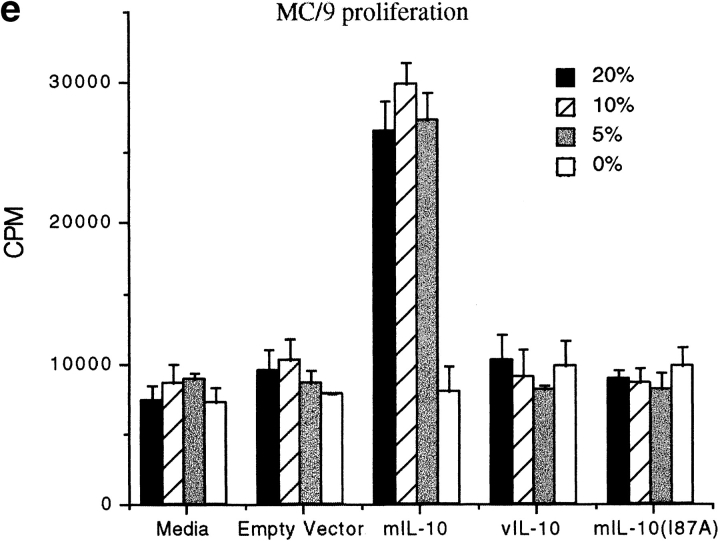
(a) Alignment of amino acid sequences of hIL-10, mIL-10 and vIL-10. Sequence and numbering based on the mature peptide without the signal sequence. Amino acid residues differing from hIL-10 are marked by boxes. (b) Chimeric IL-10 constructs made by PCR based mutagenesis. Constructs included exchange of large segments of the molecules or single amino acid changes at positions 42, 43, 87, and 89. (c) Activity of IL-10 constructs for thymocyte proliferation. Purified mIL-10/vIL-10 chimeric cDNA plasmids were transfected into COS-7 cells, supernatants were obtained after 48 h, and various concentrations were added to 2 × 105 thymocytes along with IL-2 and IL-4 for 3 d. Proliferation was assessed by 18-h [3H]thymidine incorporation. (d) Expression of various IL-10 chimeric constructs after internal labeling with [35S]methionine. Supernatants were concentrated 10-fold and analyzed on a 14% SDS-PAGE gel. (e) Activity of IL-10 constructs for MC/9 proliferation resides at amino acid 87. Various concentrations of COS supernatants were added to MC/9 cells for 24 h, and proliferation assessed by 6-h [3H]thymidine incorporation.
A review of the sequence data (Fig. 1 a) suggests that amino acids 42 and 43 (encoding QL in both murine and human, but EV in viral IL-10) and/or amino acids 87–89 (encoding IKA in human, IKE in murine, and AKD in viral IL-10) may be responsible for the functional differences. Constructs of mIL-10 cDNA with amino acids Q42E, L43V, I87A, and E89D were made (Fig. 1 b), and COS cell supernatants were again tested in the thymocyte assay (Fig. 1 c). The results clearly demonstrate that a single amino acid change at position 87(I→A) of the mature peptide sequence rendered mIL-10 inactive in the assay, whereas changes in amino acids 42, 43, or 89 had no effect on immunological activity. Thus, isoleucine 87 was critical for the immunostimulatory function in this assay. cIL-10 and vIL-10 are also discordant for their proliferative effects on mast cells: cIL-10, but not vIL-10, is a potent stimulator of mast cell proliferation, including the murine MC/9 mast cell line 18 24. To validate the thymocyte assay data and conclusions, we assayed the chimeric IL-10s in the MC/9 assay and obtained an identical immunological profile (Fig. 1 e).
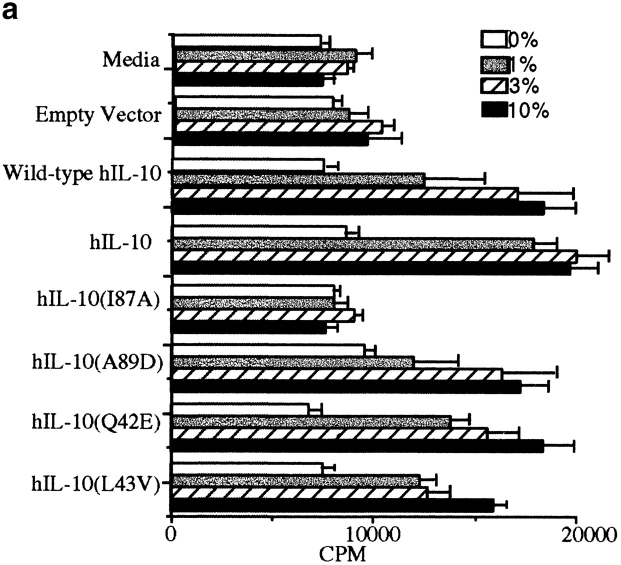
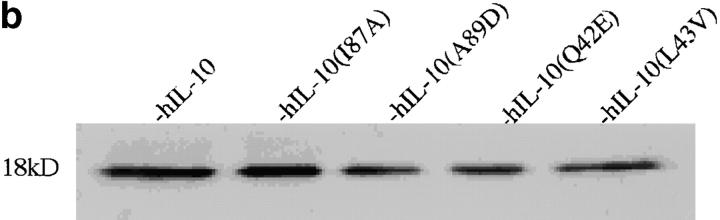
Given the close homology between murine and human IL-10s, it was important to determine if the identical changes could alter the activity of hIL-10. The appropriate constructs were cloned and tested in the MC/9 proliferation assay (Fig. 2 a). The results again demonstrated that the amino acid at position 87, but not at positions 42, 43, or 89, determined the biological properties of the cytokine, and the myc-his–tagged protein had biological activities comparable to wild-type hIL-10. Thus, a change in amino acid residue 87 of hIL-10 rendered it inactive in the MC/9 assay. Similar results were obtained with hIL-10 constructs in the murine thymocyte assay (not shown). Western blotting of the supernatants (Fig. 2 b) showed equivalent amounts of expressed protein, demonstrating again that the single amino acid change had not artifactually altered protein translocation, stability, transport, or secretion.
Figure 2.
(a) Single mutation in amino acid 87 of hIL-10 prevents MC/9 proliferation. (b) Western blotting of hIL-10 constructs shows equivalent amounts of proteins in COS-7 cell supernatants. (c) Single mutation in amino acid 87 of vIL-10 enhances MC/9 proliferation.
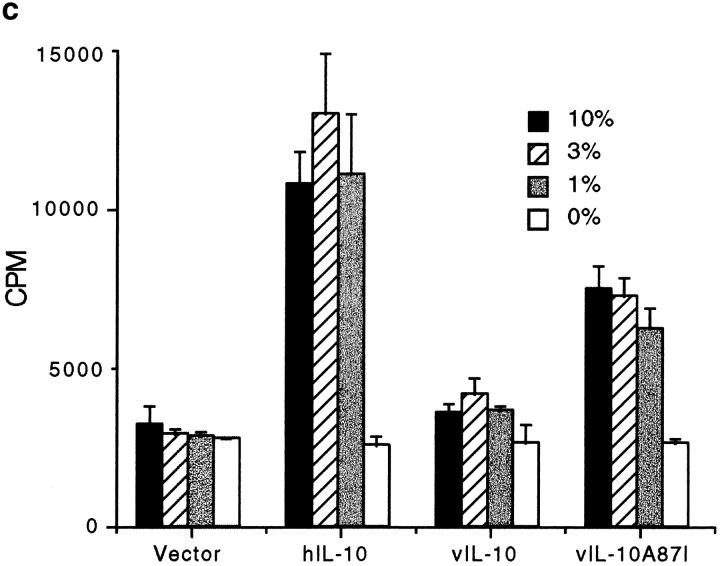
To determine if the alanine to isoleucine substitution at position 87 of vIL-10 was sufficient to allow it to acquire proliferative activity, the vIL-10(A87I) mutation was made and tested on MC/9 cells. The results show that although vIL-10(A87I) was not as active as hIL-10 in this assay, the single residue change significantly enhanced the proliferative capacity of vIL-10 (Fig. 2 c), demonstrating that isoleucine 87 was critical in determining the immunostimulatory activity of this cytokine. Although the protein content of the COS cell supernatants was not quantitated for this experiment, quantitative assays in Fig. 5 d show identical results.
Figure 5.
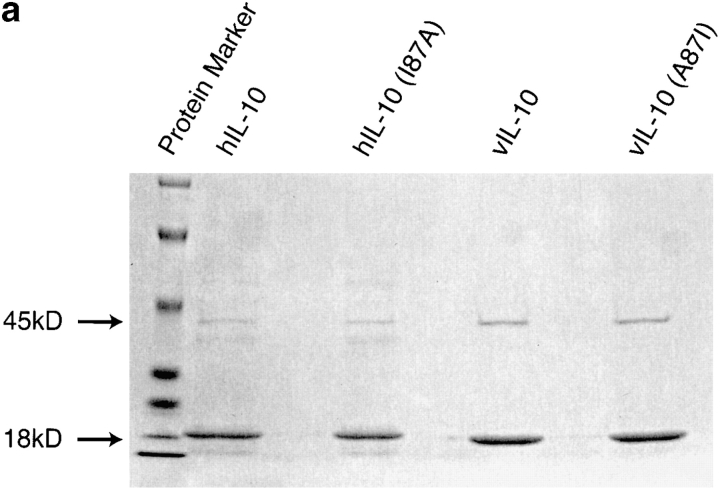
(a) Expression and purification of various IL-10s. Silver-stained SDS-PAGE of purified recombinant hIL-10, hIL-10(I87A), vIL-10, and vIL-10(A87I). IL-10 is at 18 kD; maltose binding protein (MBP) is at 45 kD. (b) A quantitative comparison of various constructs binding to mIL-10R was obtained in competitive displacement experiments. Ba/F3-mIL-10R1 cells were incubated in 100 pM 125I–hIL-10 in the presence of varying amounts of unlabeled IL-10 constructs. (c) Specific activities of various IL-10 constructs on Ba/F3-mIL-10R1 cell proliferation. (d) Specific activities of various IL-10 constructs on MC/9 cell proliferation.
Isoleucine to Alanine Substitution Does Not Alter Immunosuppressive Activities.
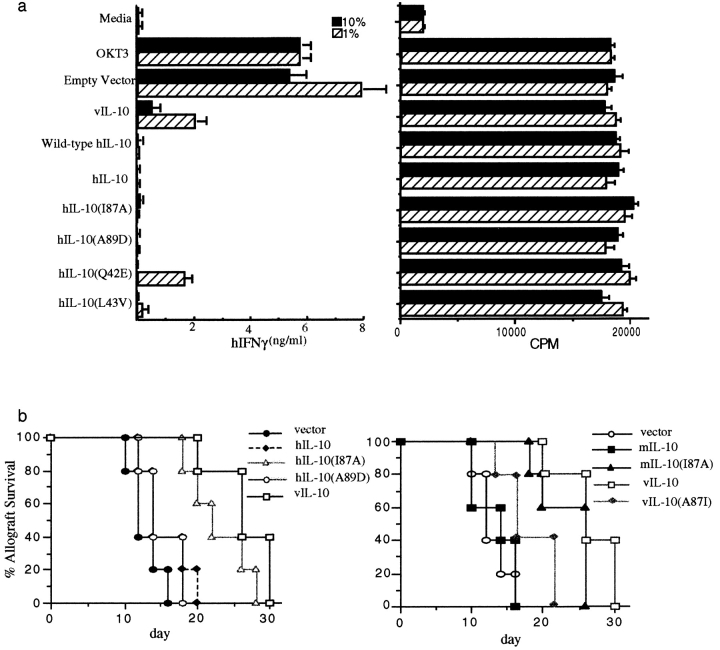
Both hIL-10 and vIL-10 inhibit IFN-γ synthesis by activated human PBMCs 18. To determine if the various constructs retained both biological and immunosuppressive activity, human PBMCs were isolated and stimulated with anti-CD3 mAb (OKT3) in the presence of COS supernatants containing various IL-10 constructs. The results in Fig. 3 a show that the various hIL-10 and vIL-10 constructs all suppressed IFN-γ production to a similar degree. Thus, suppression is the dominant effect in this assay, regardless of peptide structure. However, the constructs did not inhibit the OKT3-driven proliferative response, demonstrating that the peptides were not toxic to the cells (Fig. 3 b). mIL-10 constructs (which do not bind hIL-10R1 [36]) did not suppress IFN-γ production by human PBMCs (not shown). Together, these results demonstrate that the constructs have retained biological activity, immunosuppressive activity, and receptor species specificity.
Figure 3.
(a) IL-10 inhibits IFN-γ production but not proliferation of PBMCs. PBMCs were purified from healthy donors, and 2 × 105 cells per well were incubated with soluble OKT3 (0.1 μg/ml) plus various concentrations of IL-10 COS supernatants for 72 h. IFN γ production was measured by two-antibody capture ELISA (left), and proliferation was measured by 18-h [3H]thymidine incorporation (right). (b) A single mutation in cIL-10 allows prolongation of cardiac allograft survival. Donor neonatal C57BL/6 mouse hearts were directly injected with 0.31 μg of various IL-10 plasmid DNA constructs along with 10 μg dendrimer G5-EDA, and transplanted into CBA/J recipients. Survival of cardiac allografts was followed with EKG monitoring every other day. Only vIL-10, hIL-10(I87A), and mIL-10(I87A) prolonged cardiac allograft survival (P < 0.01 for each compared with vector).
It has been shown that cIL-10 can be immunosuppressive or immunostimulatory in vivo, whereas vIL-10 is almost exclusively immunosuppressive 26 27. Our previous report showed that rejection of mouse heart allografts was substantially delayed when the grafts were induced to express vIL-10, but not mIL-10 27. To determine if the various IL-10 constructs could suppress the alloantigen response and prolong cardiac allograft survival, plasmid DNA encoding several of the different constructs, under the control of the CMVie promoter 28, was transferred into murine cardiac allografts and the effects on graft survival were determined. The results (Fig. 3c and Fig. d) were consistent with those of the in vitro assays and our previous publication 27. vIL-10, as well as cIL-10 constructs in which amino acid 87 had been changed from isoleucine to alanine, all prolonged graft survival, whereas hIL-10, mIL-10, vIL-10(A87I), and hIL-10(A89D) did not prolong allograft survival. These results confirm the finding that a reciprocal exchange of a single amino acid can determine the spectrum of immunosuppressive or immunostimulatory activities of this cytokine. Presumably, the translation products of the IL-10 constructs interacted in vivo with T cells, B cells, APCs, and other cell types in this in vivo assay, and graft survival represents the integrated response of these diverse cell types. The results reveal that immunosuppression in vivo was associated with the alanine substitution, and that isoleucine-associated immunostimulation was dominant over the immunosuppressive activity of the peptide. This stands in contrast to the results of the IFN-γ assay in which all constructs are immunosuppressive. This likely reflects the more complex cellular interactions in vivo compared with the in vitro assay.
Isoleucine to Alanine Substitution Does Not Alter Receptor Binding Specificity and STAT Activation.
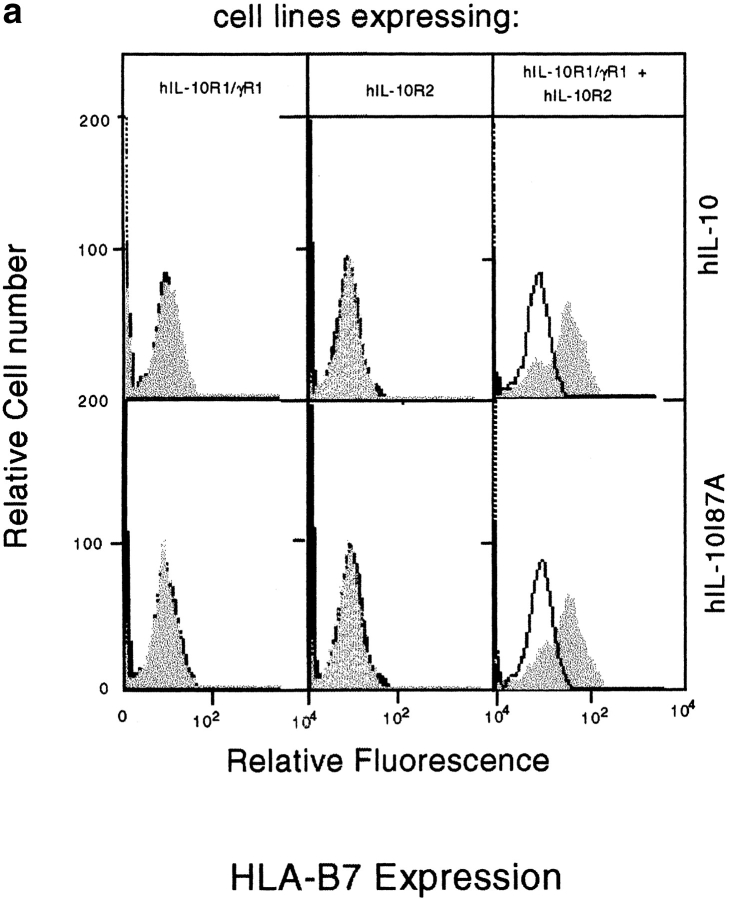
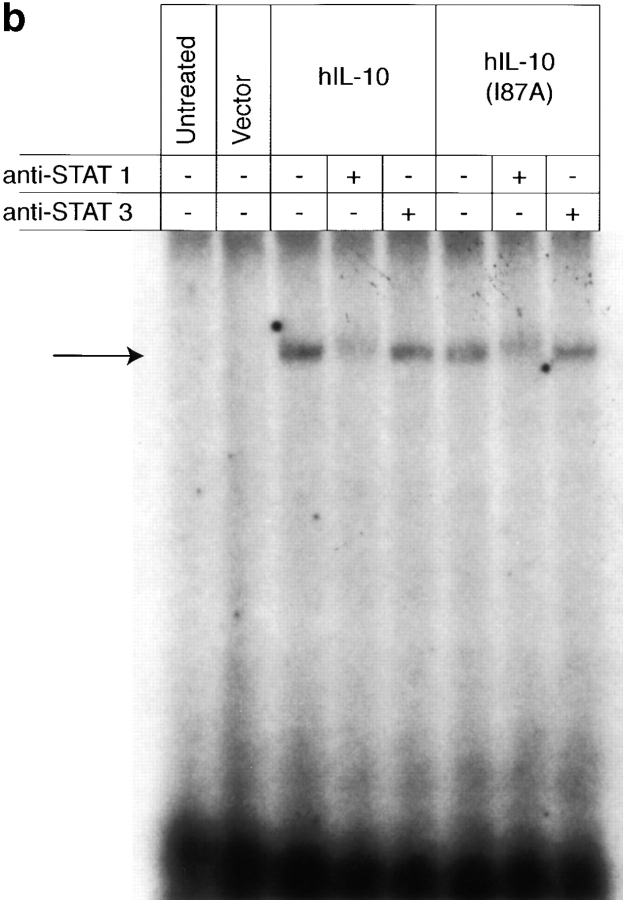
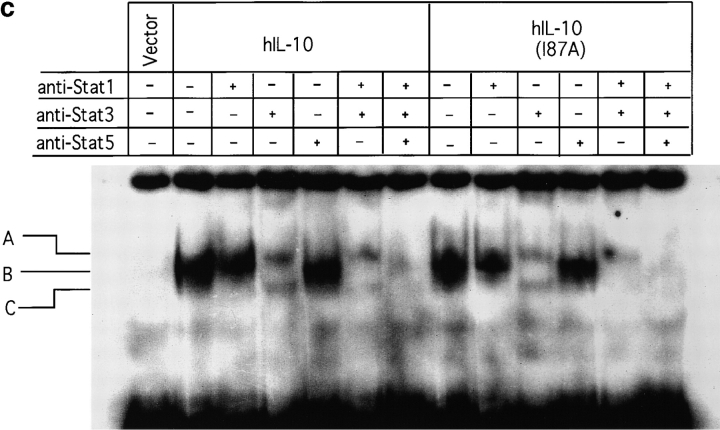
The activities of IL-10 are mediated through a cell surface receptor complex consisting of a ligand binding chain (IL-10R1) and a signal transducing accessory chain (IL-10R2) 33 39 40. Although IL-10R2 alone does not bind IL-10, only cells expressing both IL-10R1 and IL-10R2 can mediate IL-10 signal transduction 33. To determine if the single amino acid change affected the receptor binding specificity of IL-10, we stimulated 16-9 CHO cells with IL-10. These cells had been stably transfected to express an HLA-B7 reporter gene along with hIL-10R2 alone; the hIL-10R1 chimeric receptor, in which the transmembrane and intracellular domains of the hIFN-γR1 chain were substituted for the transmembrane and intracellular domains of the hIL-10R1 chain; or both receptors. We previously showed that hIL-10 stimulates IFN-γ–like responses only in those cells that express both receptors, inducing both MHC class I antigen expression and Stat1 activation 33. Fig. 4 a shows that both hIL-10 and hIL-10(I87A) induced identical MHC class I expression only in cells expressing both receptors, demonstrating that both hIL-10 and hIL-10(I87A) require IL-10R1 and IL-10R2 to mediate signal transduction, and that the single amino acid change did not alter peptide receptor binding specificity. An EMSA was performed with the same cells to determine STAT activation. The results show the comparable formation of DNA-binding complexes in both hIL-10– and hIL-10(I87A)–treated cells, and these complexes were supershifted with anti-Stat1 but not anti-Stat3 antibodies (Fig. 4 b). To explore if this result can be extended to more physiological IL-10–responsive murine cells, the IL-3–dependent murine pro-B cell Ba/F3 line expressing mIL-10R1 was also used. This line proliferates in a concordant fashion to both cIL-10 and vIL-10 29 30 31, and it was shown previously that Stat1, Stat3, and Stat5 are activated upon cIL-10 treatment in Ba/F3-mIL-10R1 cells 34. Fig. 4 c shows that both hIL-10 and hIL-10(I87A) induced Stat1, Stat3, and Stat5 activation. These results further demonstrate that the alanine substitution did not alter the receptor binding specificity and Janus kinase (JAK)-STAT signal transduction pathway activation in these particular cell lines that respond in a concordant fashion to cIL-10 and vIL-10.
Figure 4.
(a) Induction of MHC class I antigen expression by IL-10. The 16-9 CHO cell lines expressing an HLA-B7 reporter construct and hIL-10R1/hIFN-γR1 chimera and/or hIL-10R2 were exposed to control or COS cell supernatants (10%) containing hIL-10 or hIL-10(I87A) for 72 h and stained for HLA-B7. Open histograms represent negative controls; shaded histograms represent treated groups. (b) Induction of Stat1 activation by IL-10. The 16-9 cells expressing hIL-10R1/hIFN-γR1 chimera plus hIL-10R2 were exposed to COS supernatants (100%) for 15 min, the cells were lysed, and nuclear extracts were obtained for binding to 32P-labeled 22-bp IFN-γ activation sequence (GAS) from human IFN-γ regulatory factor 1 (hIRF-1). Only anti-Stat1 but not anti-Stat3 caused supershifting of the complex. (c) Induction of Stat1, Stat3, and Stat5 activation by IL-10. Ba/F3-mIL-10R1 cells were exposed to COS supernatants (100%) for 15 min, and the EMSAs were performed using a 32P-labeled 22-bp IFN-γ response region (GRR) probe.
The Single Amino Acid at Position 87 Determines Binding Affinity to IL-10 Receptors.
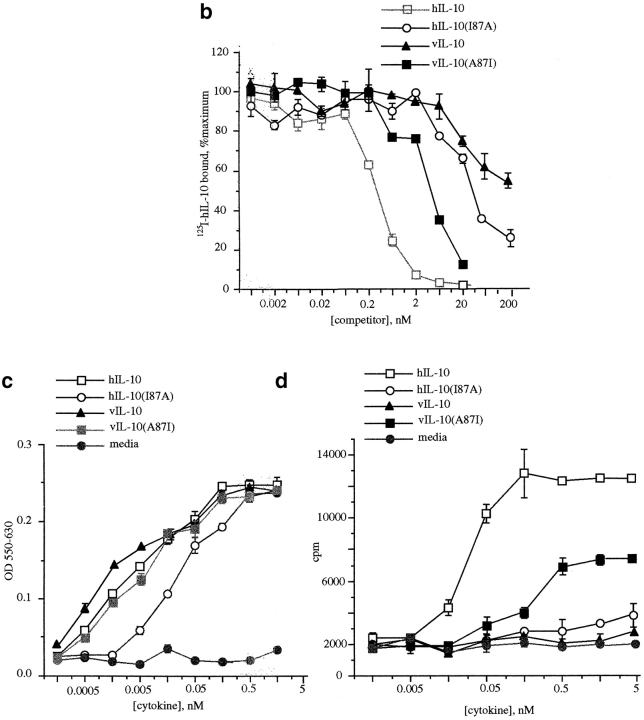
The hIL-10, hIL-10(I87A), vIL-10, and vIL-10(A87I) peptides were expressed in E. coli as described in Materials and Methods. The two-step purification of various IL-10 constructs expressed in bacteria yielded homogeneous proteins as judged by silver-stained SDS-PAGE (Fig. 5 a). No biologically functional differences between wild-type hIL-10 produced in our laboratory and commercially obtained E. coli–expressed hIL-10 were detected in the different assays performed (data not shown). A quantitative comparison of hIL-10, hIL-10(I87A), vIL-10, and vIL-10(A87I) binding to Ba/F3-mIL-10R1 cells was obtained in competitive displacement experiments (Fig. 5 b). Cells were incubated with a constant concentration of 125I-labeled hIL-10 in the presence of varying amounts of unlabeled IL-10 constructs. hIL-10 was an effective competitor for 125I-labeled hIL-10 binding; the effective 50% inhibitory concentration value was ∼200 pM. The single amino substitution at position 87(I→A) dramatically reduced its receptor binding ability; the effective 50% inhibitory concentration value for hIL-10(I87A) was at least 100-fold higher than for hIL-10, i.e., ∼20 nM. vIL-10 was a very weak competitor for 125I-labeled hIL-10 binding; however, the alanine to isoleucine substitution at position 87 dramatically enhanced its receptor binding ability, and the effective 50% inhibitory concentration value for vIL-10(A87I) was ∼6 nM, compared with at least 200 nM for vIL-10. This suggests that other residues also contribute to differences in the affinity of cIL-10 and vIL-10 for the receptor.
We compared the abilities of varying doses of hIL-10, hIL-10(I87A), vIL-10, and vIL-10(A87I) to stimulate short-term proliferation of Ba/F3-mIL-10R1 cells in order to calculate specific activities (Fig. 5 c). hIL-10 induced proliferation with a specific activity of 200 U/nM, and the single substitution at position 87(I→A) reduced its activity only ∼10-fold to 20 U/nM, despite being 100-fold less effective in receptor binding. Unexpectedly, and in contrast, vIL-10 stimulated Ba/F3-mIL-10R1 cells with a specific activity of 300 U/nM, which was higher than that of hIL-10, despite seriously impaired binding of vIL-10 compared with hIL-10 to the IL-10 receptor. Furthermore, the single amino acid substitution at position 87 of vIL-10 (A→I) slightly reduced the specific activity compared with wild-type vIL-10 (200 U/nM), despite enhanced receptor binding affinity. Although some of the apparent differences in the specific activities of these peptides could be due to some impurities or misfolding of our bacterial-derived products, the observation remains that there is a significant dichotomy between affinity and specific activity. Thus, biological responses and specific activities did not strictly correlate with ligand affinity, a phenomenon reported previously for this ligand 31, and commensurate with the results in Fig. 4b and Fig. c, in which STAT activation was not quantitatively or qualitatively altered despite changes in ligand structure and affinity.
We also compared the abilities of hIL-10, hIL-10(I87A), vIL-10, and vIL-10(A87I) to stimulate short-term proliferation in the discordant MC/9 cell assay. Fig. 5 d shows that the specific activity of hIL-10 on MC/9 cells was 20 U/nM, and therefore 10-fold less active than observed for Ba/F3-mIL-10R1 cells. Neither hIL-10(I87A) nor vIL-10 induced MC/9 proliferation over the range of concentrations tested. Thus, their low receptor affinity abrogated any specific activity in this cell line. The single amino acid substitution at position 87 of vIL-10 (A→I) enabled it to induce MC/9 proliferation. It had a specific activity of 3 U/nM, and a reduced plateau compared with hIL-10, showing that vIL-10(A87I) is a partial agonist for MC/9 cells. This correlates with its reduced receptor affinity (Fig. 5 b). Thus, in this discordant MC/9 assay (Fig. 5 d), specific activities correlated well with relative affinities, whereas in the concordant Ba/F3-mIL-10R1 assay (Fig. 5 c), specific activities did not correlate with affinity.
Discussion
These results demonstrate that a single amino acid change at position 87 of cIL-10, corresponding to the vIL-10 sequence, alters its receptor binding affinity and spectrum of biological activities. Since all of the constructs retained both immunostimulatory activity in Ba/F3-mIL-10R1 cells and immunosuppressive activity in the IFN-γ assay, the single amino acid mutations did not alter the stability of the IL-10 molecules and destroy biological function. Since reciprocal residue exchanges in cIL-10 and vIL-10 resulted in predictable receptor binding activities and biological responses, the results are unlikely to be an artifact of the system. The preservation of species specificity and the requirement for both receptor subunits for biological responses show that specific receptor binding and activation by ligand have been maintained. These results reveal separable functions for IL-10, and a new, unexpected mechanism for the regulation of IL-10 stimulatory and suppressive activities. The selective modulation of vIL-10 receptor interactions and subsequent functional spectrum by EBV may have provided a useful strategy for the virus to skew host immune responses and promote pathogenesis.
It is important to determine how this conservative amino acid change affects both the structure and function of the peptide. Structural comparisons show that the most significant differences in the amino acid sequences of viral and cellular IL-10 are found at the NH2 terminus (Fig. 1 a). Likewise, the crystal structures of hIL-10 and vIL-10 demonstrate six α-helices within each subunit of a homodimer with close topological resemblance to IFN-γ 35 36 37 38, and a significant difference in the conformation of the NH2-terminal coil of vIL-10 compared with hIL-10. The loop between α-helices A and B also differs significantly between hIL-10 and vIL-10. These differences, which are in the region of the greatest linear amino acid sequence divergence between vIL-10 and hIL-10, could account for the immunological differences between the molecules. Additional modeling comparisons to other members of the IFN family predict that the A–B loop, A helix, B helix, D helix, and F′ helix may interact with the receptor and be important for biological responses 37 39. Recently, Gesser et al. 41 performed in vitro biological activity tests on nonapeptides corresponding to the COOH and NH2 termini of hIL-10. Their results suggested that the COOH-terminal peptide possesses immunoinhibitory, whereas the NH2-terminal peptide possesses immunostimulatory, activity 41. However, the report did not show receptor binding, receptor specificity, or the biological effects of reciprocal exchanges of amino acids. Furthermore, the homologies among human, murine, and viral peptides in these regions do not correlate with the difference between cIL-10 versus vIL-10 activities. Our present results do not confirm their findings or the prediction of modeling studies 35 36 37 38. Instead, by using mIL-10/vIL-10 hybrids, we showed that the central region of cIL-10 is mainly responsible for its stimulatory activity on both murine thymocytes and mast cells. Further mutagenesis determined that the single amino acid isoleucine at position 87 is critical for receptor binding and stimulatory responses. Conversely, mutating vIL-10 from alanine to isoleucine at the corresponding position can convert vIL-10 to a partial agonist with immunostimulatory activities. Position 87 lies in the distal aspect of the loop between helices C and D and is in a region of highly conserved tertiary structure 37 39. It was unexpected that a single, conservative amino acid change could be responsible for the majority of the immunostimulatory activity of the peptide; and computer modeling also suggested this single amino acid change in cIL-10 did not alter the structural conformation (our unpublished data). Our present results imply that amino acid at position 87 likely interacts with a critical element of the IL-10 receptor complex, and IL-10 mutants change their biological responses by affecting their receptor binding affinities.
The IL-10R complex consists of two chains. IL-10R1 is a cell surface receptor with a single transmembrane domain and is a member of the class II cytokine receptor family. hIL-10R1 is species specific and does not bind mIL-10, whereas mIL-10R1 binds both mIL-10 and hIL-10. Neither hIL-10R1 nor mIL-10R1 binds vIL-10 well. Previous reports 31 and our present data show that vIL-10 bound to IL-10R1 with at least 1,000-fold less affinity than hIL-10 (Fig. 5 b). Isoleucine to alanine substitution of hIL-10 reduced its binding affinity at least 100-fold, whereas alanine to isoleucine substitution enhanced vIL-10 binding affinity by at least 100-fold. Despite these differences in affinities, the specific activities of hIL-10, hIL-10(I87A), vIL-10, and vIL-10(A87I) were similar in the Ba/F3-mIL-10R1 cells (Fig. 4 c and 5 c) and the CHO cells (Fig. 4 b). In contrast, the specific activity of hIL-10 in MC/9 cells is 10-fold lower than in Ba/F3-mIL-10R1 cells, hIL-10(I87A) and vIL-10 do not have any discernible activity on this line, and vIL-10(A87I) is a partial agonist for MC/9 cells. These latter findings correlate well with receptor affinity. The issue is to understand why significant differences in receptor affinity do or do not correlate with the specific activity of the various peptides in bioassays on particular cell lines. The lack of strict correlation between affinity and specific activity might be due to the amount of IL-10R expressed on the cells, the ratio of IL-10R1 to IL-10R2 on a cell, and/or the precise signal transduction mechanisms used by a particular cell line. The data in Fig. 4 show that the single mutation at position 87 of hIL-10 did not affect JAK-STAT signaling in two different concordant cell lines (CHO and Ba/F3). However, preliminary data (unpublished) suggest that in discordant cell lines, JAK-STAT activation is markedly sensitive to the position 87 amino acid. This finding suggests that a membrane-proximal event integrates ligand affinity and specific activity responses. The exact structural basis for this unusual finding may also relate to dynamic rather than equilibrium measurements of receptor–ligand interactions. Thus, on and off rates or receptor internalization may be more revealing than affinity. It is possible that other ligands for the IL-10R complex not yet discovered could account for some of our observations. Likewise, there could also be other receptor chains that remain undescribed at this time. However, there are constraints on these speculative possibilities. Thus, while IL-10R2 alone does not bind IL-10, it is a critical component of the receptor complex since only cells expressing both IL-10R1 and IL-10R2 can mediate IL-10 signal transduction (33; and Fig. 4 a). CRF2-4 (IL-10R2) knockout mice have characteristics similar to IL-10 knockout mice and develop chronic colitis 40, suggesting that IL-10R2 is essential for IL-10–mediated immunosuppressive effects, that IL-10R1 does not function on its own, and that possible alternative R2 chains do not mediate immunosuppression.
Acknowledgments
We wish to thank Dr. Kevin Moore for the gift of Ba/F3-mIL-10R1 cell line and for helpful discussions.
This work was supported by National Institutes of Health grants AI42840, AG15434, and UM-MAC P60 AR20557, and by the Baxter Extramural Grant Program.
Footnotes
Abbreviations used in this paper: CHO, Chinese hamster ovary; cIL-10, cellular IL-10; EMSA, electrophoretic mobility shift assay; h, human; hIL-10(I87A), the isoleucine to alanine substitution at position 87 of hIL-10; JAK, Janus kinase; m, mouse; STAT, signal transducer and activator of transcription; vIL-10, viral IL-10; vIL-10(A87I), the alanine to isoleucine substitution at position 87 of vIL-10.
References
- Fiorentino D.F., Bond M., Mosmann T. Two types of mouse helper T cells. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J. Exp. Med. 1989;170:2081–2095. doi: 10.1084/jem.170.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentino D.F., Zlotnik A., Vieira P., Mosmann T.R., Howard M., Moore K.W., O'Garra A. IL-10 acts on the antigen-presenting cell to inhibit cytokine production by Th1 cells. J. Immunol. 1991;146:3444–3451. [PubMed] [Google Scholar]
- de Waal Malefyt R., Abrams J., Bennett B., Figdor C.G., de Vries J.E. Interleukin 10 (IL-10) inhibits cytokine synthesis by human monocytesan autoregulatory role of IL-10 produced by monocytes. J. Exp. Med. 1991;174:1209–1220. doi: 10.1084/jem.174.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan C., Vodovotz Y., Nathan C. Macrophage deactivation by interleukin 10. J. Exp. Med. 1991;174:1549–1555. doi: 10.1084/jem.174.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentino D.F., Zlotnik A, Mosmann T.R., Howard M., O'Garra A. IL-10 inhibits cytokine production by activated macrophages. J. Immunol. 1991;147:3815–3822. [PubMed] [Google Scholar]
- D'Andrea A., Aste-Amezaga M., Valiante N.M., Ma X., Kubin M., Trinchieri G. Interleukin 10 (IL-10) inhibits human lymphocyte interferon γ-production by suppressing natural killer cell stimulatory factor/IL-12 synthesis in accessory cells. J. Exp. Med. 1993;178:1041–1048. doi: 10.1084/jem.178.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing K.M., Morelon E., Legendre C., De Pauw L., LeBeaut A., Grint P., Maniscalki M., Ickx B., Vereerstraeten P., Chatenoud L. A pilot trial of recombinant human interleukin-10 in kidney transplant induction therapy. Transplantation. 1997;64:999–1006. doi: 10.1097/00007890-199710150-00012. [DOI] [PubMed] [Google Scholar]
- Howard M., Muchamuel T., Andrade S., Menon S. Interleukin 10 protects mice from lethal endotoxemia. J. Exp. Med. 1993;177:1205–1208. doi: 10.1084/jem.177.4.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajkrt D., van der Poll T., Levi M., Cutler D.L., Affrime M.B., van den Ende A., ten Cate J.W., van Deventer S.J.H. Interleukin-10 inhibits activation of coagulation and fibrinolysis during human endotoxemia. Blood. 1997;89:2701–2705. [PubMed] [Google Scholar]
- Katsikis P.D., Chu C.-Q., Brennan F.M., Maini R.N., Feldmann M. Immunoregulatory role of interleukin 10 in rheumatoid arthritis. J. Exp. Med. 1994;179:1517–1527. doi: 10.1084/jem.179.5.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignon-Godefroy K., Rott O., Brazillet M.-P., Charreire J. Curative and protective effects of IL-10 in experimental autoimmune thyroiditis (EAT) J. Immunol. 1995;154:6634–6643. [PubMed] [Google Scholar]
- Kasama T., Strieter R.M., Lukacs N., Lincoln P.M., Burdick M.D. Interleukin-10 expression and chemokine regulation during the evolution of murine type II collagen–induced arthritis. J. Clin. Invest. 1995;95:2868–2876. doi: 10.1172/JCI117993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daheshia M., Kuklin N., Kanangat S., Manickan E., Rouse B.T. Suppression of ongoing ocular inflammatory disease by topical administration of plasmid DNA IL-10. J. Immunol. 1997;159:1945–1952. [PubMed] [Google Scholar]
- Rennick D.M., Fort M.M., Davidson N.J. Studies with IL-10−/− micean overview. J. Leukoc. Biol. 1997;61:389–396. doi: 10.1002/jlb.61.4.389. [DOI] [PubMed] [Google Scholar]
- van Deventer S.J.H., Elson C.O., Fedorak R.N. Multiple doses of intravenous interleukin 10 in steroid-refractory Crohn's disease. Gastroenterology. 1997;113:383–389. doi: 10.1053/gast.1997.v113.pm9247454. [DOI] [PubMed] [Google Scholar]
- Moore K.W., Vieira P., Fiorentino D.F., Trounstine M.L., Khan T.A., Mosmann T.R. Homology of cytokine synthesis inhibitory factor (IL-10) to the Epstein-Barr virus gene BCRFI. Science. 1990;248:1230–1234. doi: 10.1126/science.2161559. [DOI] [PubMed] [Google Scholar]
- Hsu D.-H., de Waal Malefyt R., Fiorentino D., Dang M.-N., Vieira P., de Vries J., Spits H., Mosmann T.R., Moore K.W. Expression of interleukin-10 activity by Epstein-Barr virus protein BCRF1. Science. 1990;250:830–832. doi: 10.1126/science.2173142. [DOI] [PubMed] [Google Scholar]
- Vieira P., de Waal-Malefyt R., Dang M.-N., Johnson K.E., Kastelein R., Fiorentino D.F., de Vries J.E., Roncarolo M.-G., Mosmann T.R., Moore K.W. Isolation and expression of human cytokine synthesis inhibitory factor (CSIF/IL-10) cDNA cloneshomology to Epstein-Barr virus open reading frame BCRF1. Proc. Natl. Acad. Sci. 1991;USA. 88:1172–1176. doi: 10.1073/pnas.88.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Waal Malefyt R., Haanen J., Spits H., Roncarolo M.-G., te Velde A., Figdor C.G., Johnson K., Kastelein R., Yssel H., de Vries J.E. Interleukin 10 (IL-10) and viral IL-10 strongly reduce antigen-specific human T cell proliferation by diminishing the antigen-presenting capacity of monocytes via downregulation of class II major histocompatibility complex expression. J. Exp. Med. 1991;174:915–924. doi: 10.1084/jem.174.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Prete G., De Carli M., Almerigigna F., Giudizi M.G., Biagiotti R., Romagnani S. Human IL-10 is produced by both type 1 helper (Th1) and type 2 helper (Th2) T cell clones and inhibits their antigen-specific proliferation and cytokine production. J. Immunol. 1993;150:353–360. [PubMed] [Google Scholar]
- Punnonen J., de Waal Malefyt R., van Vlasselaer P., Gauchat J.-F., de Vries J.E. IL-10 and viral IL-10 prevent IL-4-induced IgE synthesis by inhibiting the accessory cell function of monocytes. J. Immunol. 1993;151:1280–1289. [PubMed] [Google Scholar]
- MacNeil I.A., Suda T., Moore K.W., Mosmann T.R., Zlotnik A. IL-10, a novel growth cofactor for mature and immature T cells. J. Immunol. 1990;145:4167–4173. [PubMed] [Google Scholar]
- Fei N.F., Castle B.E., Barrett R., Kastelein R., Dang W., Mosmann T.R., Moore K.W., Howard M. Interleukin 10, a novel B cell stimulatory factorunresponsiveness of X chromosome–linked immunodeficiency B cells. J. Exp. Med. 1990;172:1625–1631. doi: 10.1084/jem.172.6.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Snipes L.A., Dhar V., Bond M.W., Mosmann T.R., Moore K.W., Rennick D.M. Interleukin 10a novel stimulatory factor for mast cells and their progenitors. J. Exp. Med. 1991;173:507–510. doi: 10.1084/jem.173.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F., Garcia E., Defrance T., Peronne C., Vezzio N., Hsu D.-H., Kastelein R., Moore K.W., Banchereau J. Interleukin 10 is a potent growth and differentiation factor for activated human B lymphocytes. Proc. Natl. Acad. Sci. USA. 1992;89:1890–1893. doi: 10.1073/pnas.89.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Tahara H., Narula S., Moore K.W., Robbins P.D., Lotze M.T. Viral interleukin 10 (IL-10), the human herpes virus 4 cellular IL-10 homologue, induces local anergy to allogeneic and syngeneic tumors. J. Exp. Med. 1995;182:477–486. doi: 10.1084/jem.182.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L., Chavin K.D., Ding Y., Tahara H., Favaro J.P., Woodward J., Lin J., Suzuki T., Robbins P.D., Lotze M.T., Bromberg J.S. Retroviral mediated transfer of viral IL-10 gene prolongs murine cardiac allograft survival. J. Immunol. 1996;156:2316–2323. [PubMed] [Google Scholar]
- Philip R., Brunette E., Kilinski L., Murugesh D., McNally M.A., Ucar K., Rosenblatt J., Okarma T.B., Lebkowski J.S. Efficient and sustained gene expression in primary T lymphocytes and primary and cultured tumor cells mediated by adeno-associated virus plasmid DNA complexed to cationic liposomes. Mol. Cell. Biol. 1994;14:2411–2418. doi: 10.1128/mcb.14.4.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Kang S.-H., Heidenreich O., Li Q., Nerenberg M. Rapid PCR method for site-directed mutagenesis on double-stranded plasmid DNA. Biotechniques. 1996;20:44–46. doi: 10.2144/96201bm09. [DOI] [PubMed] [Google Scholar]
- Ho A.S.-Y., Liu Y., Khan T.A., Hsu D.-H., Bazen J.F., Moore K.W. A receptor for interleukin-10 is related to interferon receptors. Proc. Natl. Acad. Sci. USA. 1993;90:11267–11271. doi: 10.1073/pnas.90.23.11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., de Waal Malefyt R., Briere F., Parham C., Bridon J.-M., Banchereau J., Moore K.W., Xu J. The EBV IL-10 homologue is a selective agonist with impaired binding to the IL-10 receptor. J. Immunol. 1997;158:604–613. [PubMed] [Google Scholar]
- Qin L, Pahud D.R., Ding Y., Bielinska A.U., Kukowska-Latallo J.F., Baker J.R., Bromberg J.S. Efficient transfer of genes into murine cardiac grafts by Starburst polyamidoamine dendrimers. Hum. Gene Ther. 1998;9:553–560. doi: 10.1089/hum.1998.9.4-553. [DOI] [PubMed] [Google Scholar]
- Kotenko S.V., Krause C.D., Izotova L.S., Pollack B.P., Wu W., Pestka S. Identification and functional characterization of a second chain of the interleukin-10 receptor complex. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:5894–5903. doi: 10.1093/emboj/16.19.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Nordt R.M., Riley J.K., Greenlund A.C., Moore K.W., Darnell J.E., Schreiber R.D. Stat3 recruitment by two distinct ligand-induced, tyrosine-phosphorylated docking sites in the interleukin-10 receptor intracellular domain. J. Biol. Chem. 1996;271:27954–27961. doi: 10.1074/jbc.271.44.27954. [DOI] [PubMed] [Google Scholar]
- Zdanov A., Schalk-Hihi C., Gustchina A., Tsang M., Weatherbee J., Wlodawer A. Crystal structure of interleukin-10 reveals the functional dimer with an unexpected topological similarity to interferon γ. Structure. 1995;3:591–601. doi: 10.1016/s0969-2126(01)00193-9. [DOI] [PubMed] [Google Scholar]
- Walter M.R., Nagabhushan T.L. Crystal structure of interleukin 10 reveals an interferon γ-like fold. Biochemistry. 1995;34:12118–12125. doi: 10.1021/bi00038a004. [DOI] [PubMed] [Google Scholar]
- Zdanov A., Schalk-Hihi C., Wlodawer A. Crystal structure of human interleukin-10 at 1.6 Å resolution and model of a complex with its soluble receptor. Protein Sci. 1996;5:1955–1962. doi: 10.1002/pro.5560051001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdanov A., Schalk-Hihi C., Menon S., Moore K.W., Wlodawer A. Crystal structure of Epstein-Barr virus protein BCRF1, a homolog of cellular interleukin-10. J. Mol. Biol. 1997;268:460–467. doi: 10.1006/jmbi.1997.0990. [DOI] [PubMed] [Google Scholar]
- Liu Y., Wei H.-Y., Ho A.S.-Y., Malefyt R., Moore K.W. Expression cloning and characterization of a human interleukin-10 receptor. J. Immunol. 1994;152:1821–1829. [PubMed] [Google Scholar]
- Spencer S.D., Di Marco F., Hooley J., Pitts-Meek S., Bauer M., Ryan A.M., Sordat B., Gibbs V.C., Aguet M. The orphan receptor CRF2-4 is an essential subunit of the interleukin 10 receptor. J. Exp. Med. 1998;187:571–578. doi: 10.1084/jem.187.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesser B., Leffers H., Jinquan T., Vestergaard C., Kirstein N., Sindet-Pedersen S., Jensen S.L., Thestrup-Pedersen K., Larsen C.G. Identification of functional domains on human interleukin 10. Proc. Natl. Acad. Sci. USA. 1997;94:14620–14625. doi: 10.1073/pnas.94.26.14620. [DOI] [PMC free article] [PubMed] [Google Scholar]