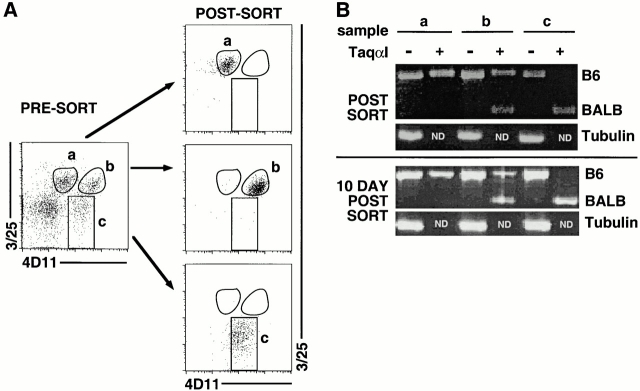
Figure 3.
Cell sorting and identification of Ly49G2 populations. Pooled, freshly isolated nylon wool nonadherent spleen cells from (B6 × BALB.B)F1 mice were stained with NK1.1, 3/25, and 4D11. (A) Three populations of NK1.1+ cells were separated by cell sorting: (a) 3/25+4D11low; (b) 3/25+4D11high; and (c) 3/25−4D11+. As tested by postsort analysis, the populations were each of >90% purity. (B) The Ly49G2 alleles expressed by each population were determined by RT-PCR with Ly49G2 specific primers. Before electrophoresis, PCR products were digested with Taqα1, a diagnostic restriction enzyme that cleaves only the Ly49G2BALB cDNA. RT-PCR analysis was performed on cells immediately postsort (top) and after in vitro expansion for 10 d in the presence of IL-2 (bottom). Because the tubulin PCRs were tested only without digestion, the Taqα1 lanes are empty (ND). The experiment was repeated with NK cells from class I–deficient mice with identical results.