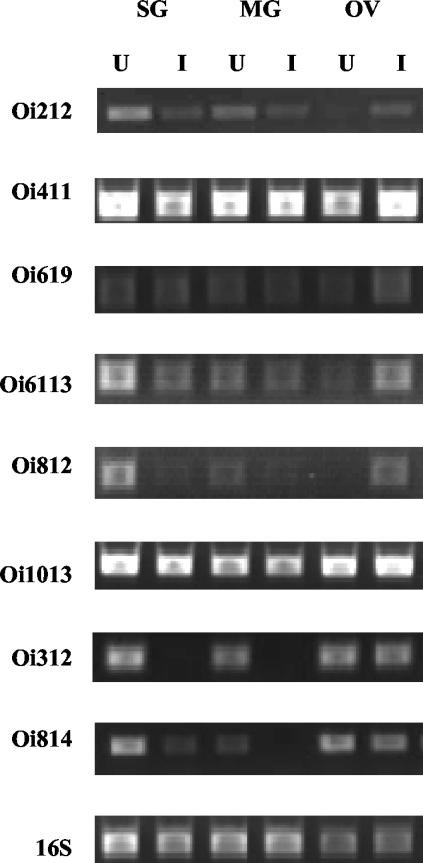
FIG. 2.
Semiquantitative RT-PCR analysis of D. variabilis cDNAs that were identified as differentially regulated in response to rickettsial infection. RNA from uninfected and Rickettsia-infected salivary glands (SG), midguts (MG), and ovaries (OV) was used to generate first-strand cDNA. By using clone-specific primers (Table 1) and cDNA as the template, comparative PCR analysis of expression was carried out. Above each lane, U and I represent uninfected and Rickettsia-infected samples, respectively. D. variabilis 16S primers served as a positive load control.