Figure 4.
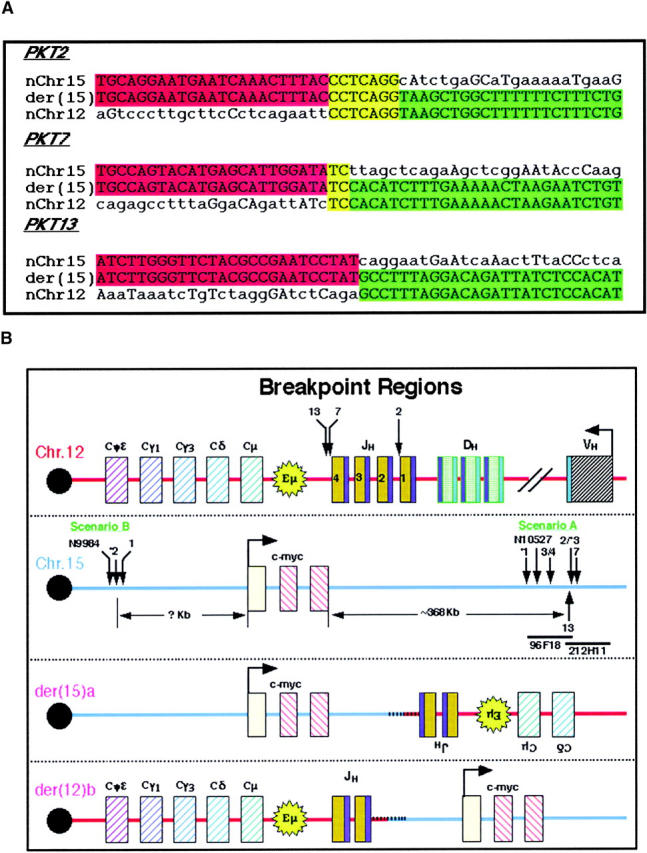
Sequence analysis and localization of fusion points and rearrangements in the IgH Ca and c-myc loci. (A) The fusion points of three Ku80−/−p53−/− pro-B cell lymphomas (PKT no. 2, 7, and 13) were cloned by digestion circularization PCR [der(15)a]. Sequence analysis of the three breakpoints from scenario A demonstrate microhomology (yellow) at the junction of chromosomes 12 (red) and 15 (green) in two cases (PKT2 and PKT7), but none in tumor PKT13. Homologous bases outside of this region are also indicated in uppercase letters. (B) Sequence analysis implied that the breakpoint was the V(D)J recombination signal sequence (RSS; blue) flanking IgH J1 in PKT2 and IgH J4 in PKT7 and PKT13 (top panel); however, exonuclease digestion of the cleaved ends resulted in loss of 45, 208, and 189 bp, respectively, from the original site of the RAG-mediated DSB to the fusion point (arrows). Approximate localization of the breakpoints on chromosome 15 (second panel) was determined by FISH hybridization with two BAC clones in this region (96F18 and 212H11) as illustrated in Fig. 5. Tumors 1–4, 7, and 13 are from Ku80−/−p53−/− mice, tumors *1–*3 are from Ku70−/−p53−/− mice and tumors N9984 and N10527 are from Ku80−/−p53−/−RAD54−/− mice. The resulting der(15)a and der(12)b chromosomes are illustrated in the bottom panels. In scenario A a portion of the IgH locus is copied to chromosome 15 downstream of c-myc and in the reverse orientation. In scenario B it appears that the copying of chromosome 15 to the der(12)b initiates centromeric of the c-myc locus and continues along toward the telomere.
