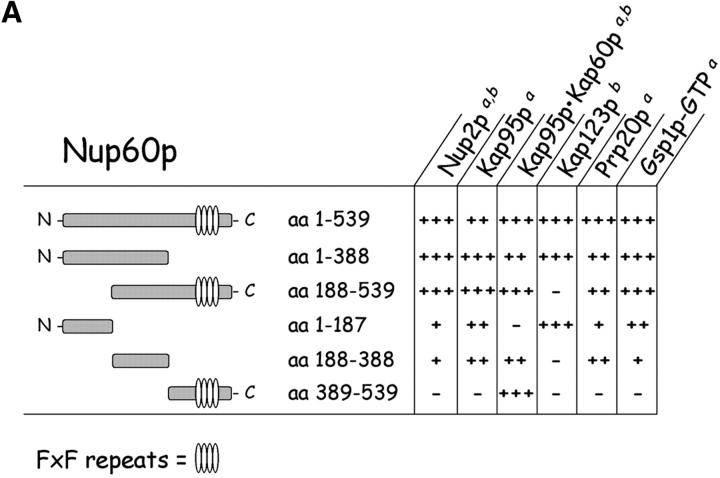
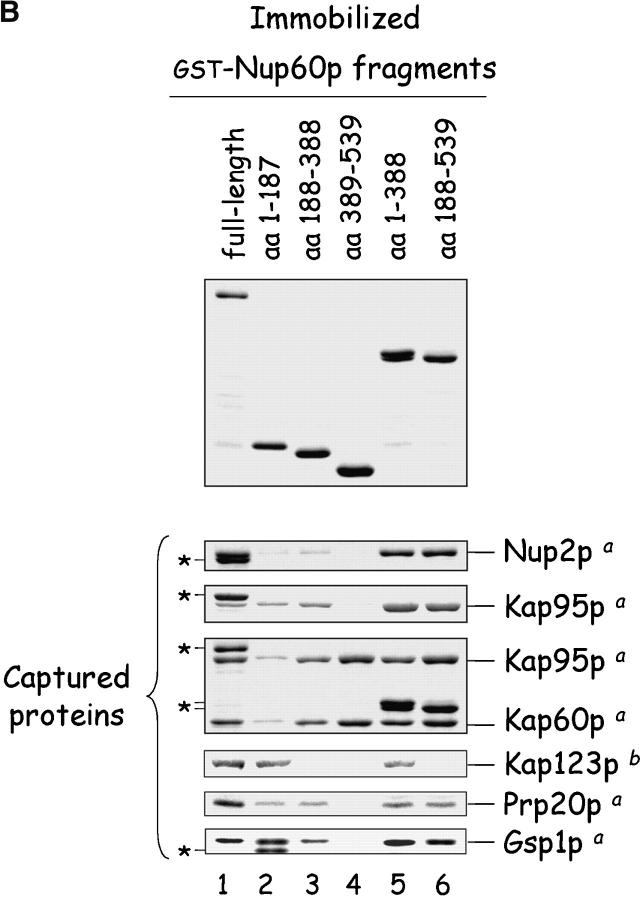
Figure 6.
Mapping of Nup, karyopherin, Prp20p, and Gsp1p–GTP binding sites on Nup60p. (A) The cartoon depicts the Nup60p fragments used in this study provides a summary of the binding interactions observed in B. The + and − designations provide a qualitative assessment of the binding avidity. The absence of detectable binding is denoted by −, whereas +, ++, and +++ represent relative degrees of binding. (B) Binding of karyopherins, Nup2p, Prp20p, and Gsp1p–GTP to various Nup60p fragments. Each GST–Nup60p fragment (1 μg each) was immobilized on beads and incubated with purified Nup2p, Kap95p, Kap95p–Kap60p heterodimers, Prp20p, or Gsp1p–GTP (1 μg each), as indicated. After 1 h at 4°C, the beads were washed and bound proteins were collected, resolved by SDS-PAGE, and visualized with Coomassie blue stain. Alternatively, each GST–Nup60p fragment (5 μg each) was immobilized on beads and incubated with 10 mg of yeast extract for 2 h at 4°C. After washing the beads, bound proteins were eluted with 1 M NaCl, collected by precipitation with trichloroacetic acid and deoxycholate, resolved by SDS-PAGE, and stained with Coomassie blue. Asterisks designate GST–Nup60p fragments used as bait. The superscripts “a” and “b” denote the source of proteins used in the experiments: “a” marks cases where purified recombinant proteins where used, and “b” marks cases where yeast extracts were used. Note the shift in binding site selection of Kap95p in the presence and absence of Kap60p.