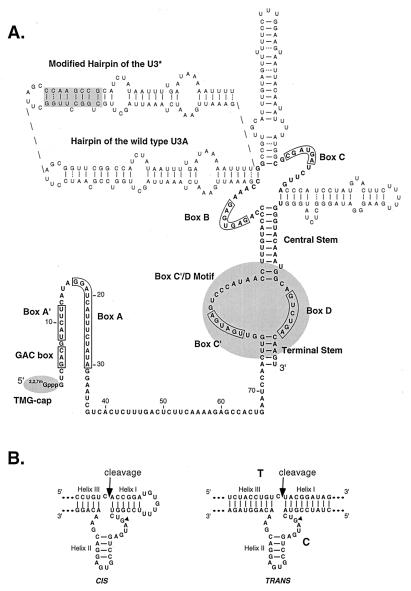
Figure 1.
Structure of the U3 snoRNA-hammerhead ribozyme (snorbozyme) chimeras featured in the present study. (A) Map of S. cerevisiae U3 (22). The yeast U3 molecule consists of (i) a 5′ region with little or no secondary folding; (ii) a single-stranded hinge region, and (iii) a highly structured 3′ domain. The 5′ and 3′ ends are protected from exonucleolytic damage by a trimethyl guanosine (TMG) cap and one or more proteins complexed with the box C′/D motif. Key elements incorporated into the design of the snorbozyme system are emphasized. The 5′ domain contains highly conserved box elements GAC, A′, and A, which are implicated in direct interaction with pre-rRNA. The box C′/D motif is a nucleolar localization signal and provides metabolic stability. The in vivo studies were carried out with cells expressing wild-type U3, which is essential for growth, and one or two U3 variants containing the experimental sequences. One variant (U3*) contains a unique hybridization tag created by modifying the sequence of one of the helical stems (upper left). The second is a functional “mini-U3” molecule (U3del), in which all hairpin domains are absent. The ribozyme and target sequences were introduced into the 5′ region of the U3 host molecules in place of conserved boxes A′ and A and a portion of the hinge segment. (B) Structures of the hammerhead ribozyme elements featured, in cis and trans configurations (8, 25). A G nucleotide essential for ribozyme cleavage is marked with a filled triangle. Inactive snorbozymes containing a C at this position were used in control experiments. The target and catalytic sequences in the trans configuration are labeled T and C, respectively.