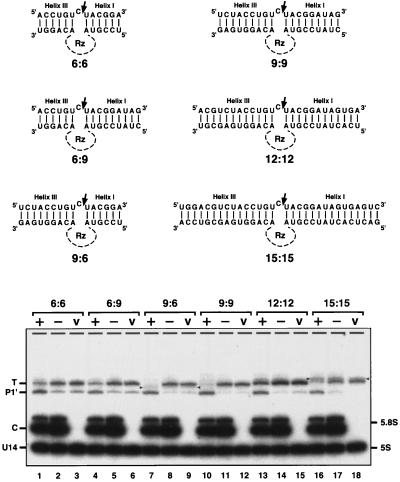
Figure 5.
Effect of flanking complementarity on trans-acting ribozyme cleavage activity. (A) A set of substrates able to form different numbers of base pairs with the initial ribozyme molecule was prepared. The relevant target region is on top, paired with the corresponding ribozyme moiety on the bottom (the catalytic core of the ribozyme Rz is omitted for simplicity). The trans-acting ribozyme complex examined earlier is the 9:9 pair. (B) Efficiency of cleavage was assessed by using Northern blot analysis. Each substrate RNA was coproduced with an active (+) or inactive (−) ribozyme molecule or in the presence of an empty host vector (V). Variable amounts of truncated substrates are evident in the negative control samples. The yield of these molecules reflects differences in the 5′ sequences of the substrates and the growth stage of the cells used in the analysis (more is present at later stages of growth). These products are presumed to arise from random degradation of the substrate 5′ end. The origin of the truncated substrate variant in lane 17 is not clear. Accumulation of this product might reflect a nonspecific cleavage process that depends on the inactive ribozyme. Alternatively, cleavage might be mediated by the “inactive” ribozyme, which may have low residual activity that is magnified in this particular, high-affinity context. The RNA species marked with triangles (lanes 7–9 and 16–18) most likely originate from alternative folding of target molecules. Similar effects have been observed previously with internally modified U2 and U3 RNAs (22, 26). Interestingly, these target molecules are completely resistant to ribozyme cleavage.