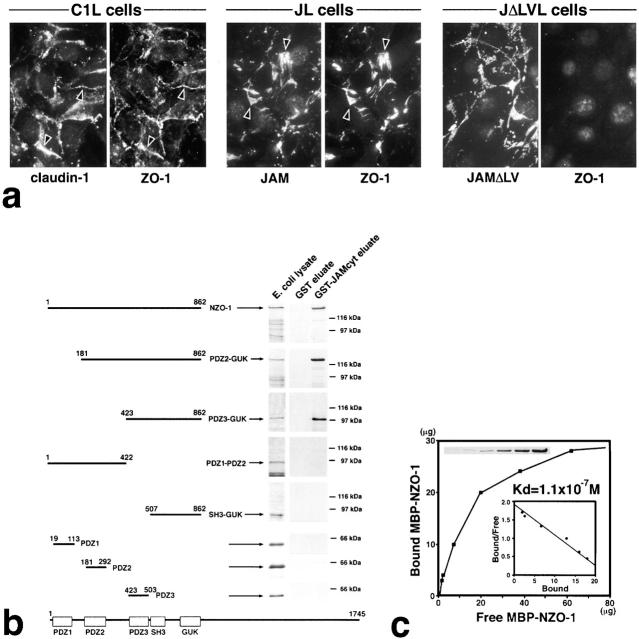
Figure 3.
Interaction between JAM and ZO-1. (a) Recruitment of endogenous ZO-1 to cell–cell adhesion sites in L transfectants. C1L cells, JL cells, or JAM lacking its COOH-terminal –LV (JΔLVL cells) were double stained. Claudin-1, JAM, and JAMΔLV were all concentrated at cell–cell adhesion sites. Claudin-1 and JAM, but not JAMΔLV, recruited ZO-1 to cell–cell contact sites (arrowheads). (b) Eight distinct portions of ZO-1 were produced as recombinant fusion proteins with maltose-binding protein (MBP) in E. coli. Their crude lysates containing recombinant proteins (E. coli lysate) were mixed with beads conjugated with GST or GST fusion protein with the cytoplasmic domain of JAM (GST-JAMcyt). Bound proteins were then eluted from GST-conjugated beads (GST eluate) or GST-JAMcyt–conjugated beads (GST-JAMcyt eluate), and each eluate was subjected to SDS-PAGE followed by Coomassie brilliant blue staining. Among eight types of MBP fusion proteins, only MBP–NZO-1, MBP-PDZ2-GUK, and MBP-PDZ3-GUK were bound to GST-JAMcyt. (c) Quantitative analysis of the binding between MBP-NZO-1 and GST-JAMcyt. Glutathione–Sepharose bead slurry containing GST-JAMcyt was incubated with E. coli lysate containing various amounts of MBP–NZO-1. The amounts of MBP–NZO-1 in the E. coli lysate and each eluate (inset) were estimated by comparing the Coomassie Brilliant blue staining intensity of bands. The binding was saturable, and Scatchard analysis (inset) indicated that the K d value was 1.1 × 10−7 M.