Figure 2.
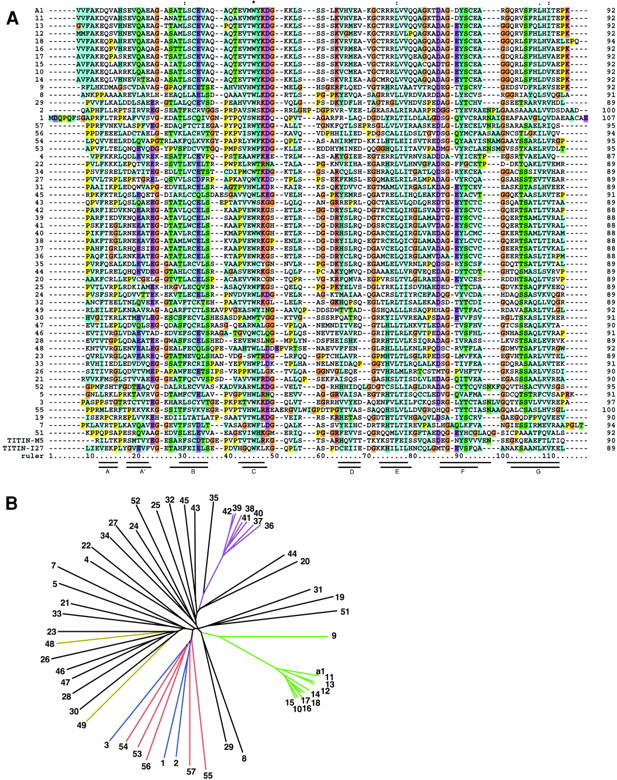
Comparison of obscurin Ig domains. (A) Multiple sequence alignment of all obscurin Ig domains using ClustalW (Higgins et al., 1996). Note the extremely high homology of some groups of adjacent domains. Also included are titin M5 and I27 domains whose structures have been solved. The seven β-strands of M5 (top) and I27 (bottom) are indicated as bars under the alignment (Pfuhl and Pastore, 1995; Improta et al., 1996). This alignment was used to generate the phylogenetic tree in B. Residue coloring: all prolines, yellow; all glycines, brown; conserved basic residues, pink; conserved acidic residues, purple; conserved hydrophobic residues, blue. (B) A phylogenetic tree of the obscurin Ig domains does not reveal any patterns of super repeats as seen in titin. In some regions, consecutive domains cluster together and are highly homologous, for example domains Ob9–15 and Ob36–42 (green and purple, respectively). This suggests a rapid expansion of the domain pattern during the evolution of the protein. The three NH2-terminal domains (blue) cluster together with the five most COOH-terminal ones (red). The titin binding domains Ob48 and Ob49 are colored yellow.
