Figure 5.
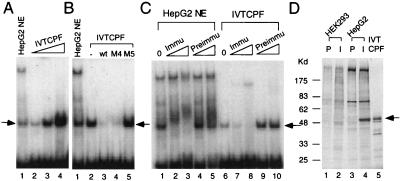
Functional similarity of in vitro-synthesized and endogenous CPF. (A) EMSAs with in vitro-synthesized CPF-1 (IVTCPF) and CL1 as the radio-labeled probe. Lanes 2–4, 0.1, 0.5 and 1.0 μl of in vitro-synthesized CPF-1 (IVTCPF), respectively, and lane 1, 1 μg of HepG2 nuclear extract (NE). The arrow indicates the CPF binding complex. (B) Competition EMSAs. EMSAs were performed as described above with the addition of unlabeled competitor oligonucleotides, including wild type (wt) and those with mutations in either the CPF binding site (M5) or a region outside of the CPF binding site (M4). A 60-fold molar excess of each competitor was used. The arrow denotes the location of the bound DNA-protein complex. NE, HepG2 nuclear extract; −, no competitor, IVTCPF, in vitro-translated CPF. (C) EMSA in the presence of CPF-specific antibodies. EMSAs were performed with HepG2 nuclear extracts and in vitro-synthesized CPF with the addition of either preimmune serum (Preimmu) or rabbit anti-CPF antibodies (Immu). Lanes 1 and 6, no antibody (0); lanes 2, 4, 7, and 9, 0.1 μl of serum; lanes 3, 5, 8, and 10, 1 μl of serum. The arrow denotes the location of the bound DNA-protein complex. (D) Immunoprecipitation of CPF in HepG2 cells. HEK293 and HepG2 cells were grown in the presence of [35S]methionine. Preimmune (P) and rabbit anti-CPF antibodies (I) then were used in immunoprecipitation experiments with HEK293 (lanes 1 and 2) and HepG2 (lanes 3 and 4) extracts. 35S-methionine-labeled in vitro-synthesized CPF (IVTCPF) was loaded into lane 5. The precipitated samples were separated by 10% SDS/PAGE, and gel was dried and exposed to x-ray film. The arrow denotes the immunoprecipitated and in vitro-synthesized CPF. Molecular size markers (in kd) are shown on the left.