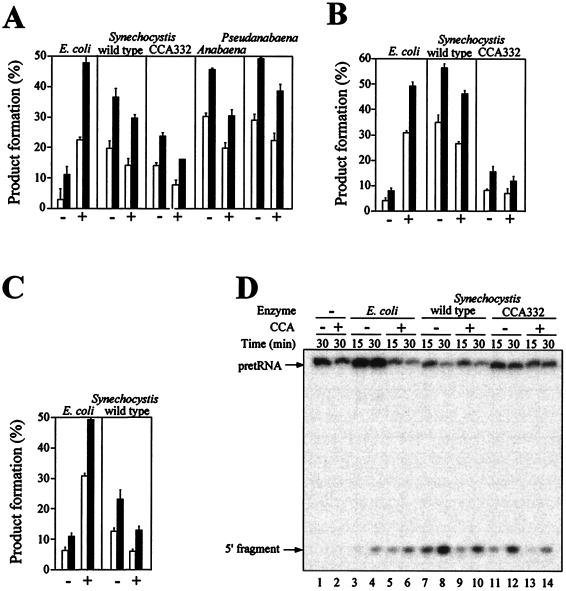
Figure 4.
Effect of 3′ terminal CCA in pGln and pGlu on pre-steady-state cleavage kinetics by cyanobacterial RNase P RNAs or holoenzymes. (A–C) Ten picomolar pre-tRNA (−) or pre-tRNACCA (+) was incubated with the indicated RNase P RNAs or reconstituted holoenzymes for 15 min (open bars) or 30 min (solid bars), and the amount of processing was determined. For the RNA-alone reactions, the concentrations used were 0.5 nM of E. coli M1 RNA and 5 nM of all the other RNase P RNAs. The E. coli holoenzyme was reconstituted with 0.1 nM M1 RNA and 1 nM C5 protein. The Synechocystis wild-type and CCA332 mutant holoenzymes were reconstituted with 2 nM RNase P RNA and 20 nM Synechocystis RNase P protein. The average ± SD of three independent experiments is shown. (A) pGln and pGlnCCA cleavage by RNase P RNA. (B) pGln and pGlnCCA cleavage by RNase P holoenzyme. (C) pGlu and pGluCCA cleavage by RNase P RNA. (D) Autoradiogram of a representative assay gel. Ten picomolar 5′ end-labeled pGln (−) or pGlnCCA (+) was incubated for 15 min (lanes 3, 5, 7, 9, 11, and 13) or 30 min (lanes 1, 2, 4, 6, 8, 10, 12, and 14) in the absence of enzyme (lanes 1 and 2) or with 0.5 nM E. coli M1 RNA (lanes 3–6), 5 nM Synechocystis RNase P RNA wild type (lanes 7–10), or 5 nM Synechocystis RNase P RNA CCA332 (lanes 11–14).