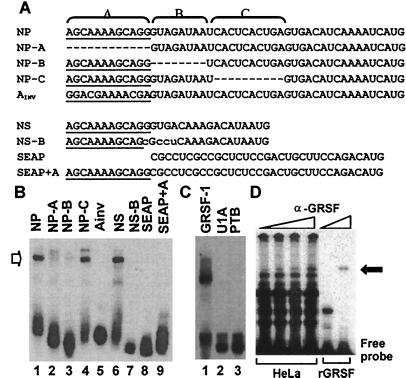
Figure 2.
GRSF-1 interacts with specific sequences within the influenza virus mRNA 5′ UTR as detected by gel mobility-shift analysis. (A) Sequences of the in vitro transcribed RNA transcripts used as probes in gel mobility-shift assays. Underlined sequences represent the conserved 12-nt sequences found on all influenza virus type A mRNAs. The 5′ UTR of NP mRNA was divided into three regions (regions A, B, and C) as depicted across the top. The name of each transcript is indicated on the left side of its sequence. AINV is a derivative of NP, in which region A is reversed. The NS 5′ UTR is shown along with a sequence of the substitution mutant NS-B (mutated bases are shown in lowercase letters). Below are shown the sequences of the SEAP 5′ UTR along with the SEAP 5′ UTR appended to region A (SEAP+A). (B) Recombinant GRSF-1 (0.050 μg) purified from E. coli was incubated in the presence of the nonspecific competitor, heparin (0.125 mg/ml), with the various probes indicated across the top, as described under Materials and Methods. (C) The RRM-containing RNA-binding purified proteins U1A and PTB (0.050 μg) were incubated with the NP 5′ UTR probe as specificity controls (for details, see B). (D) HeLa S10 extract (200 μg, Left) or recombinant GRSF-1 (rGRSF-1) (0.050 μg, Right) was incubated with the NP 5′ UTR in the presence of increasing amounts of monoclonal anti-GRSF-1 for supershift analysis. The resulting RNA–protein complexes were resolved on a native polyacrylamide gel. The GRSF-1-RNA complex (open arrow on the left) and its antibody complex (solid arrow on the right) are indicated.