Figure 3.
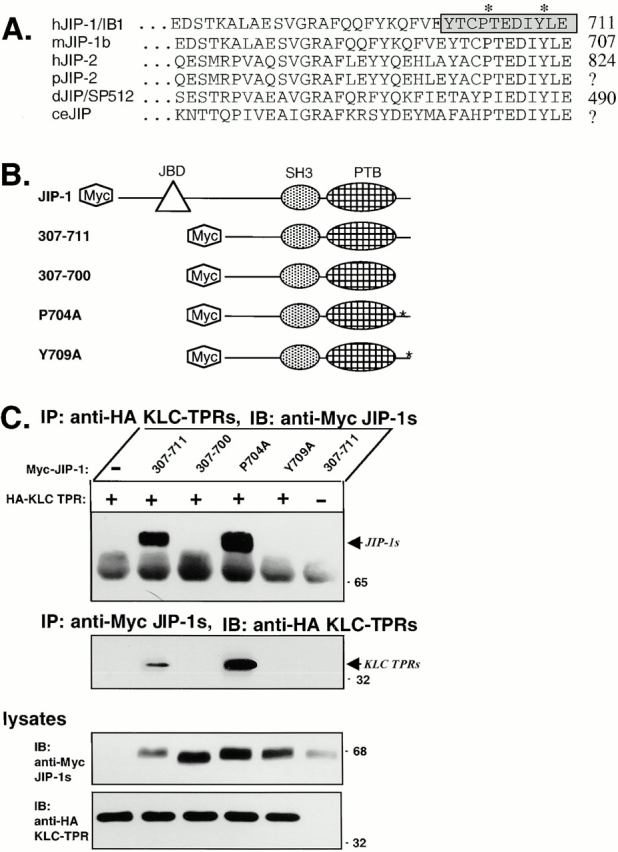
The COOH-terminal residues of JIP-1 are required for interaction with the KLC TPRs. (A) Sequence alignment of the COOH-terminal residues of JIP-1 and JIP-2 across species. The 11 residues deleted in the mutant JIP-1 (307–700) are indicated (boxed and shaded) as are the residues mutated in JIP-1 (P704A) and JIP-1 (Y709A) (*). hJIP-1/IB1, human JIP-1; mJIP-1b, mouse JIP-1b; hJIP-2, human JIP-2; pJIP-2, pig JIP-2 derived from a partial EST (accession AW312953); dJIP/SP512, Drosophila JIP-1/2; ceJIP, C. elegans JIP sequence derived from cosmid C13A10. (B) Schematic illustration of the Myc-tagged JIP-1 constructs used in this study. (C) Lysates of 293 cells expressing Myc-tagged JIP-1 (307–711), JIP-1 (307–700), JIP-1 (P704A), or JIP-1 (Y709A) together with HA-tagged KLC TPRs were immunoprecipitated (IP) for the KLC TPRs using an anti-HA mAb (top) or for the JIP-1 variants using an anti-Myc mAb (middle). Precipitates were immunoblotted (IB) for associated proteins using anti-Myc (top) or anti-HA polyclonal antibodies (middle). The ∼65-kD band in the top panel is the heavy chain of immunoglobulin. Lysates were immunoblotted to detect the expressed proteins (bottom).
