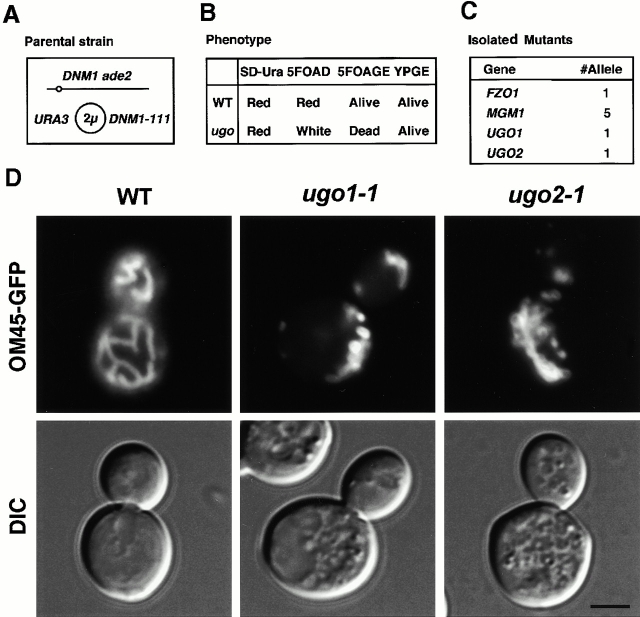
Figure 1.
Isolation of ugo mutants. (A) Strain used to isolate ugo1 and ugo2. ade2 DNM1 strains that carry plasmid pHS50, which expresses the dominant negative Dnm1-111 protein, lack Dnm1p activity. Cells that lose pHS50 contain functional Dnm1p. (B) Mutant isolation scheme. The parental strain (WT) maintains mtDNA on glucose-containing medium in the presence (SD-Ura) or absence (5FOAD) of the URA3-DNM1-111 plasmid and form red colonies due to the ade2 mutation (Reaume and Tatum 1949). ugo mutants maintain mtDNA only in the presence of the pHS50, forming red colonies on SD-Ura, but become white on 5FOAD medium, which selects for loss of the URA3-DNM1-111 plasmid (Boeke et al. 1984). Growth on glycerol and ethanol medium confirms that ugo mutants contain mtDNA in the presence of the URA3-DNM1-111 plasmid (YPGE), but that ugo mutants are inviable on 5FOA medium containing glycerol and ethanol (5FOAGE). (C) Four different genes were identified in the ugo screen. Complementation tests showed one fzo1 mutant, five mgm1 mutants, and two new mutants, ugo1 and ugo2, were isolated. (D) ugo mutants contain highly fragmented mitochondria. Wild-type (W303), ugo1-1 (YHS64), and ugo2-1 (YHS65) were grown on 5FOAD medium to select for cells that lost the URA3-DNM1-111 plasmid, pHS50, and then transformed with pKC2, which expresses GFP fused to the COOH terminus of the mitochondrial outer membrane protein, OM45p (OM45-GFP; Cerveny et al. 2001). Cells were grown to log phase in SRaf medium and examined by fluorescence microscopy. Fluorescence (OM45-GFP) and DIC images are shown. Bar, 3 μm.