Figure 1.
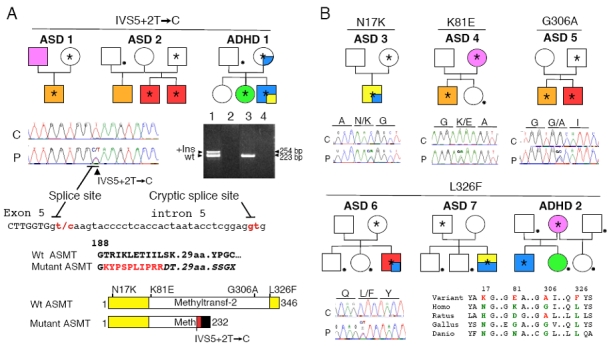
Non-synonymous ASMT variations in autism spectrum disorders (ASD) families. (a) Pedigree structure of the families carrying the splice–site mutation IVS5+2T>C; reverse transcriptase-polymerase chain reaction (RT-PCR) amplifying exons 4 to 6 of the ASMT cDNA from B lymphoblastoid cell lines of the ASD 1 proband carrying the splice– site mutation IVS5+2T > C (lane 1 +RT, lane 2 −RT) and a control (lane 3 +RT, lane 4 −RT). The insertion (+Ins) of 31 bp in the ASMT transcript originates from a cryptic donor splice–site downstream from exon 5. This insertion should lead to the additional sequence indicated in red and to a frame shift (characters in italics), causing premature truncation of the protein, lacking the methyl-transferase domain. Wt, wild-type. (b) Pedigree structure of the families carrying rare non-synonymous ASMT variations and conservation of the variant amino-acid in different species. Color codes in the pedigrees: autism with mild (orange) or severe (red) mental retardation, Asperger syndrome or high-functioning autism (yellow), attention-deficit/hyperactivity disorder ADHD (light blue) and depression (pink). The proband ASD 3 fulfilled diagnostic criteria for both high functioning autism and ADHD. The proband ASD 7 fulfilled diagnostic criteria for both Asperger syndrome and ADHD. The asterisk and the dot indicate the presence of the mutation and the absence of a DNA sample for analysis, respectively.
