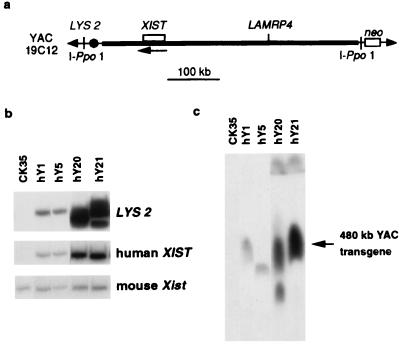
Figure 1.
Structural analysis of human XIST transgenes. (a) Human YAC 19C12 is 480 kb long (24). XIST is flanked by about 380 kb of 5′ and 70 kb of 3′ sequence. The XIST gene and LAMRP4 pseudogene are shown. The orientation of XIST transcription is indicated with an arrow. Meganuclease I-Ppo1 sites were introduced into both YAC vector arms to facilitate assessment of YAC integrity in transgenic cells (25) along with the pgk-neo-selectable marker, introduced into one arm, to enable selection for YAC uptake after lipofection into CK35 male ES cells. (b) EcoRI-digested DNA of control CK35 ES cells and transgenic clones hY1, hY5, hY20, and hY21 hybridized with probes for: LYS2, a yeast marker present in one of the YAC vector arms (Top); human XIST (6/7r in exon 1) (Middle); and mouse Xist (HF) (Bottom). Using these probes and others, transgene copy number was estimated to be 1–2 for lines hY1 and hY5, 8–10 for line hY20, and 10–14 for hY21. (c) I-Ppo1-digested DNA of transgenic clones hY1, hY5, hY20, and hY21 resolved on a pulse-field gel. Hybridization with either XIST (shown here), LAMRP4, or YAC vector probes revealed a 480-kb band corresponding to intact YAC sequences in lines hY1, hY5, hY20, and hY21. Lanes corresponding to these clones, from the same autoradiograph, have been juxtaposed. The band in hY5 appears slightly lower in size as a result of aberrant migration because of DNA-loading differences between lanes (as judged by ethidium bromide staining). In line hY20, the additional smaller fragment suggests the presence of some rearrangement in a subpopulation of the transgenic sequences.