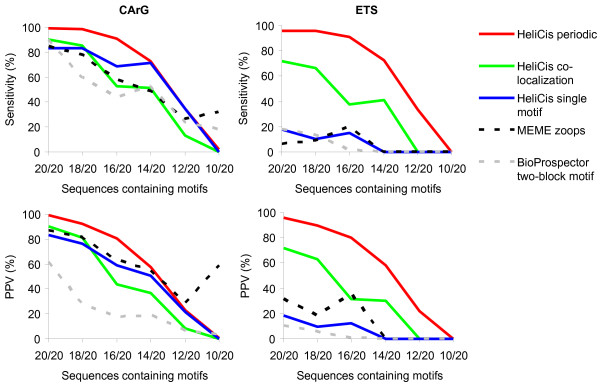
Figure 4.
Performance on synthetic sequence datasets with varying motif coverage. Datasets of 20 sequences with colocalized and periodically spaced CArG and ETS motifs were generated. The proportion of sequences containing the motifs was gradually reduced, thus making them increasingly difficult to detect. HeliCis with different settings was compared to MEME and BioProspector. The plots show sensitivity and positive predictive value (PPV = TP/(TP + FP)). Results are from 5 averaged trials.