Figure 1.
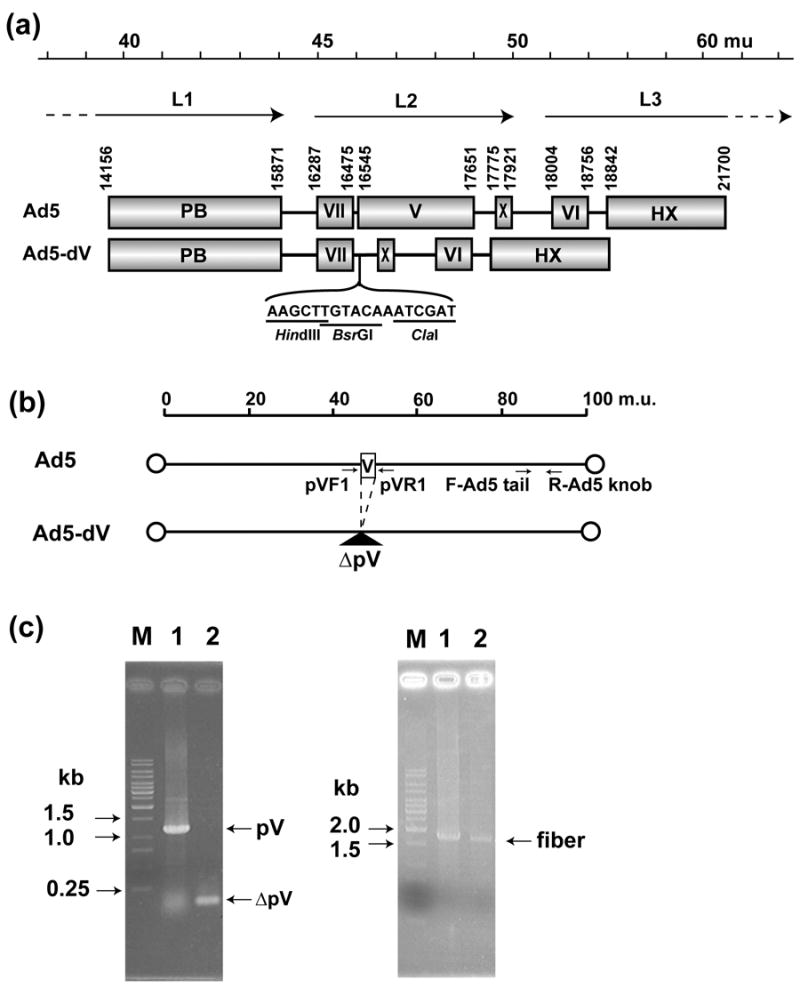
Genomic structure of the Ad5-dV and its PCR validation scheme. (a) Genomic structures of the wild type Ad5 and the pV-deletion mutant (Ad5-dV). The genome of Ad5-dV is carrying a deletion of nucleotides at position 16,544-17,650 of the wild type Ad5, thus this deletion removes the entire coding sequence of the pV gene. mu, map units; PB; penton base, VII; polypeptide VII, V; polypeptide V, X; polypeptide X (Mu), VI; polypeptide VII, HX; hexon, L1–L3; major late transcript units. (b) Schematic representation of the Ad5-dV genome PCR validation strategy. PCR primer pairs flanking pV and fiber gene sequences are indicated by arrows. pVF1, pV forward primer; pVR1, reverse pV primer; F-Ad5 tail, Ad5 tail sequence-specific forward primer; R-Ad5 knob, Ad5 knob sequence-specific reverse primer. (c) Validation of a pV deletion in the Ad5-dV genome by PCR. Left panel, PCR with pV flanking primers (pVF1 and pVR1); Right panel, PCR with fiber-specific primers (F-Ad5tail and R-Ad5knob); M, DNA size markers; Lane 1, PCR using total DNAs from wild type Ad5-infected cells as a template; Lane 2, PCR using total DNA from the Ad5-dV-infected cells as a template.
