Figure 1.
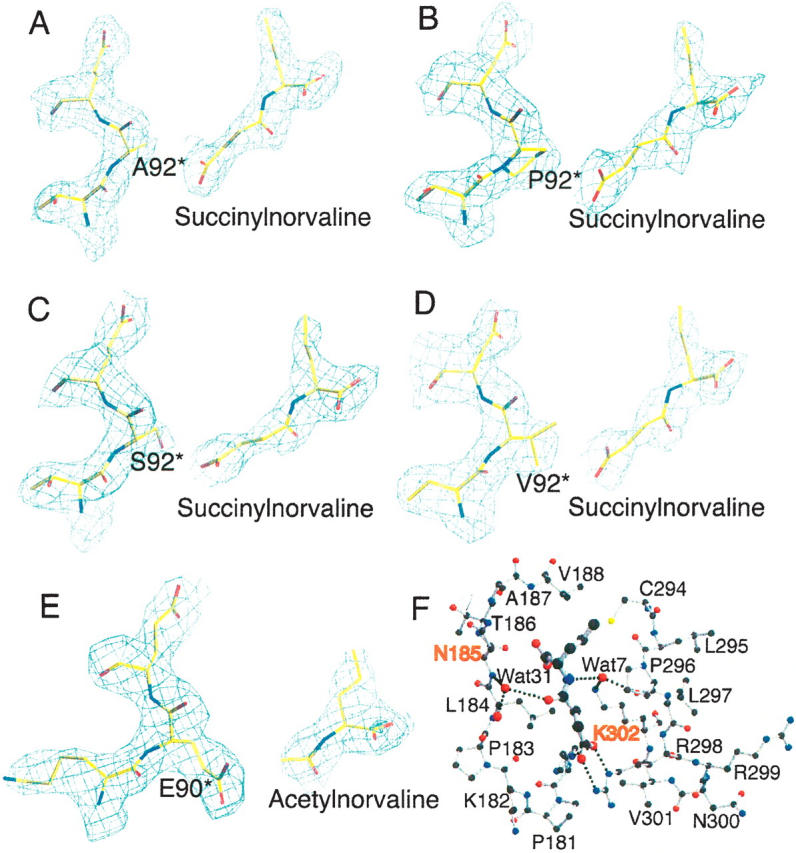
N-acetylnorvaline or N-succinylnorvaline binding of SOTCase or AOTCase mutants. (A–D) Contours of the electron density map (2Fo − Fc) (1.0 σ shown in blue cage) around N-succinylnorvaline and E92 region of X. campestris AOTCase mutants. (E) Contours of the electron density map (2Fo − Fc) (1.0 σ shown in blue cage) around N-acetylnorvaline and P90 region of B. fragilis SOTCase mutant. The final refined positions of the ligands and residues are represented as colored sticks. The residue indicated by * is from the adjacent subunit. The electron density clearly indicates the binding of substrate analogs and the expected amino acid substitutions. (F) N-succinylnorvaline binding site of X. campestris E92A mutant. SNOR is shown as thick ball-and-stick models, while the surrounding residues are shown as thin ball-and-stick models. Two water molecules, Wat7 and Wat31, form hydrogen-bonding bridges to link the ligand to protein residues. These water-mediated hydrogen-bonding networks can only be found in X. campestris AOTCase and its mutants, but not in B. fragilis SOTCase and mutants. The residues, N185 and K302 (X. campestris AOTCase numbering) shown in red, are proposed as the residues that distinguish SOTCase from AOTCase in addition to the key residue of E92.
