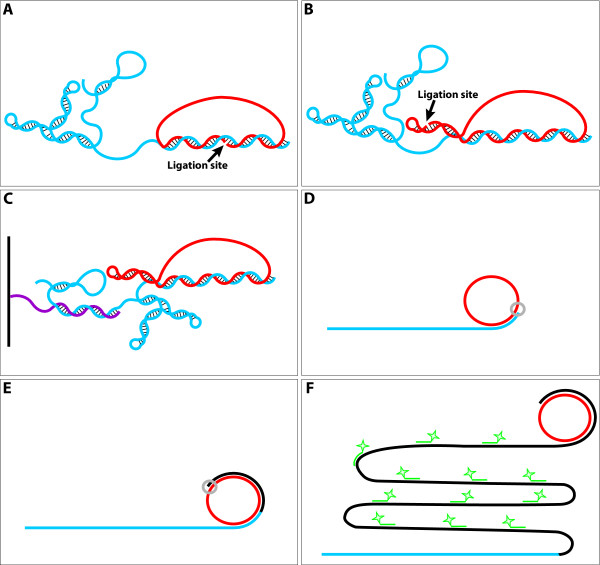
Figure 1.
Probe designs and methods for RNA detection. (A) A Padlock Probe contains two ends that are brought into close proximity through hybridization to a target RNA molecule (blue) after which they can be joined by a DNA ligase (the ligation site is indicated by an arrow). The probe also contains an intervening segment which does not hybridize to the target molecule, but completes the circle. This intervening segment can be rather freely designed in terms of length and sequence and is used as the probe identifier. Thus probes may be equipped with individual intervening segments for unique identification in multiplexed experiments. (B) A Turtle Probe consists of a target recognizing element and the identifier joined by a hairpin structure bringing the probe ends into close proximity on an internal ligation template (the ligation site is indicated with by arrow). (C) Illustration of the solid support setup where the capture oligonucleotide (purple) is covalently attached to the glass, the target RNA (blue) is hybridized to the capture oligonucleotide and the Turtle Probe (red) is hybridized to the 3'-end of the target RNA. (D-F) The target RNA (blue) provides a free 3'-end for the rolling circle reaction employing a ligated circle probe (red) (could be a Padlock- or Turtle Probe) as template for the localized DNA synthesis (grey polymerase forms black DNA). The rolling circle product, extending from the 3'-end of the target RNA, is then visualized with labeled oligonucleotide probes (green) recognizing the copies of the identifier element produced in the rolling circle reaction.