Figure 1.
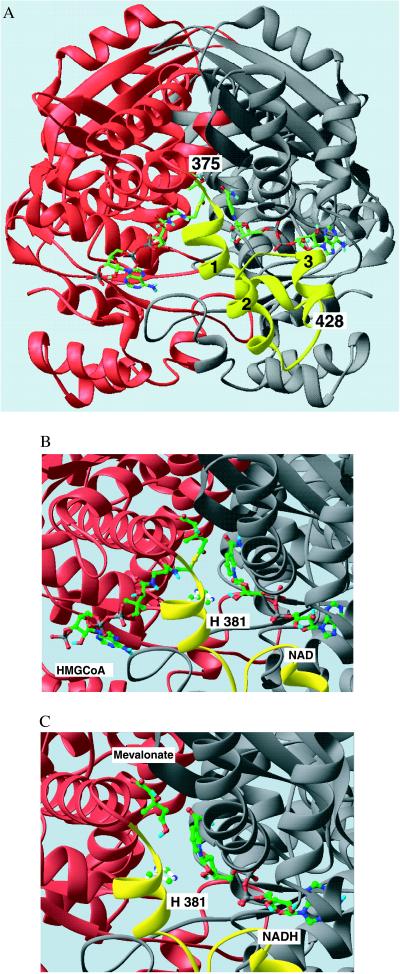
(A) Ribbon diagram of the HMG-CoA reductase dimer with HMG-CoA and NAD+ bound. The view looks directly into the active site cavity between the two monomers shown in red and gray. The flap domain, in yellow, extends from residue 375 in the red monomer in three helical segments (marked 1–3) to the C-terminal residue of the molecule, residue 428. The substrates are shown as stick models with C = green, N = blue, O = red, P = gray, and S = yellow. Both substrates bind in an extended conformation with, HMG-CoA binding to the large domain of the red monomer on the left and NAD+ binding to the small domain of the gray monomer on the right. (B and C) Close-up views of the active site in the ternary complexes of HMG-CoA reductase with HMG-CoA/NAD+ and mevalonate/ NADH, respectively. The catalytic His381 side chain in the closed flap is shown as a ball and stick model, positioned near the sulfur of the HMG-CoA in B. The similar position of the HMG moiety of HMG-CoA and of mevalonate in these complexes is readily seen. The NAD(H) nicotinamide ring in the anti/pro-S configuration lays side-by-side with the substrate, positioned for hydride transfer. This figure was prepared with the program ribbons (19).