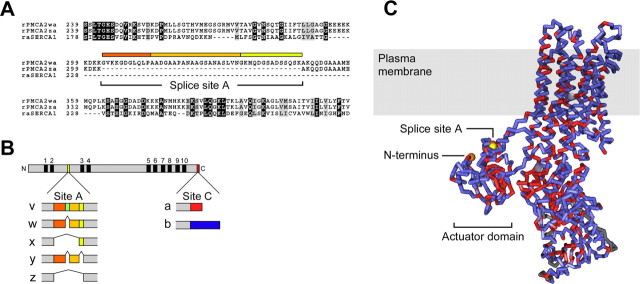
Figure 1.
Location of splice-site A in the amino acid sequence and predicted structure of PMCA2. A, Alignment of amino acid sequences of rabbit muscle SERCA with rat PMCA2 splice variants in the vicinity of splice-site A. SERCA lacks sequences corresponding to this splicing region. B, Scheme of the PMCA protein and diagram of splicing at sites A and C in frog and rat PMCA2. The PMCA is shown at the top as a gray bar; the 10 membrane-spanning domains are indicated by black boxes and numbered from 1 to 10. N, N terminus; C, C terminus. Colored boxes denote the positions where alternative splicing alters PMCA2 at sites A and C. Splicing at site A affects the first cytosolic loop between membrane-spanning domains 2 and 3; splicing at site C alters the C-terminal sequence of the pump protein. The colored boxes below the PMCA scheme indicate the amino acid insertions encoded by separate exons in different PMCA2 splice variants (labeled on the left). The colors of the boxes correspond to the colored bars above the sequence in A. Note that a unique sequence insertion in the 2v splice form of frog PMCA2 (indicated by an additional dark green box) interrupts the 2w sequence. This sequence insertion does not exist in rat (or human) PMCA2 and hence is not shown in A; frog PMCA2v is nevertheless targeted to hair bundles. C, Side view of the SERCA structure (1SU4). PMCA2 identities are red; nonidentical but aligned amino acids are blue. Gray areas do not align between the two sequences. The location of the actuator domain, N terminus (orange), and splice-site A (yellow) are indicated. The shaded box corresponds to the approximate location of the membrane bilayer. Note the proximity of the N terminus to splice-site A. The structure was generated with Cn3D version 4.1.