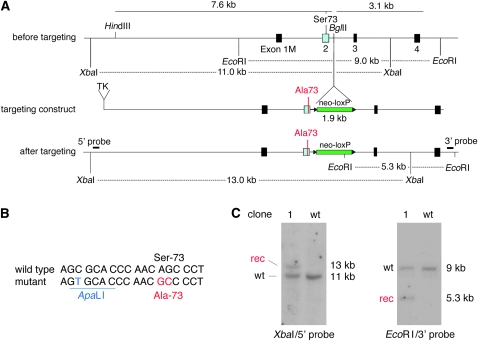
Figure 1.—
Targeting construct and analysis of a targeted ES clone. (A) Schematic of a portion of the mouse Mitf gene and targeting construct. (Top) The genomic region before targeting around exon 1M–4, the position of S73 in exon 2, the position of the BglII site where the neo-cassette will be inserted, and the position of the EcoRI and XbaI restriction sites used to identify legitimate recombination events. (Middle) The targeting construct, consisting (in the 5′–3′ direction) of a TK cassette, 7.6 kb of 5′ flanking sequence containing a modified codon, the neocassette (green) flanked by loxP sites (triangles), and 3.1 kb of 3′ flanking sequence. (Bottom) The genomic arrangement after targeting, showing the position of the diagnostic restriction fragments and the position of the probes used for Southern hybridization. The XbaI fragment in wild type is 11 kb but after targeting is increased to 13 kb because of the insertion of the neo-cassette. The EcoRI fragment in wild type is 9 kb and, after targeting, a novel EcoRI fragment of 5.3 kb is generated because of an EcoRI site in the neo-cassette. (B) Wild-type and modified sequence around S73. The mutant contains a silent ApaLI site and an AGC-to-GCC codon change, leading to a Ser-73-to-Ala change. (C) Analysis of wild-type and clone 1 ES cells by Southern hybridization. Note the expected recombinant bands after indicated restriction cuts and hybridization with the indicated probes.