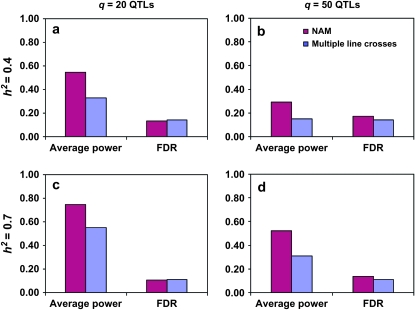
Figure 6.—
Comparison of average power and FDR for NAM analysis and traditional linkage analysis of multiple line crosses. Significance threshold was set at α = 10−7. (a) q = 20 QTL and h2 = 0.4; (b) q = 50 QTL and h2 = 0.4; (c) q = 20 QTL and h2 = 0.7; (d) q = 50 QTL and h2 = 0.7. For NAM analysis, SNP information between CPS markers was projected from founders to 5000 RILs and a true positive was counted only if the QTL locus was retained in the final model; for linkage analysis, no projection was done and a unique allele was assumed for each founder and a true positive was counted as long as the locus retained in the final model was located within the region containing a QTL.