TABLE 5.
Estimated haplotype, polygenic, total genetic (±SE), and residual (±SE) variances and heritabilities at different values of r2 between adjacent markers for the low-heritability trait
| Analysis | r2 SNP | Haplotype | Polygenic | Total geneticab | Residuala | h2 |
|---|---|---|---|---|---|---|
| Simulatedc | 0.10 | 0.90 | 0.10 | |||
| Polygenic | 0.143 | 0.873 | 0.140 | |||
| SNP1 | 0.103 | 0.016 | 0.128 | 0.144 | 0.869 | 0.141 |
| 0.120 | 0.020 | 0.109 | 0.129 | 0.872 | 0.128 | |
| 0.152 | 0.029 | 0.089 | 0.118 | 0.874 | 0.119 | |
| 0.189 | 0.043 | 0.070 | 0.113 | 0.863 | 0.116 | |
| 0.210 | 0.054 | 0.053 | 0.107 | 0.863 | 0.110 | |
| SNP2 | 0.103 | 0.021 | 0.125 | 0.146 | 0.867 | 0.143 |
| 0.120 | 0.027 | 0.108 | 0.134 | 0.868 | 0.133 | |
| 0.152 | 0.037 | 0.080 | 0.117 | 0.871 | 0.119 | |
| 0.189 | 0.053 | 0.058 | 0.111 | 0.864 | 0.113 | |
| 0.210 | 0.065 | 0.048 | 0.113 | 0.859 | 0.117 | |
| HAP_IBD2 | 0.103 | 0.036 | 0.079 | 0.114 | 0.866 | 0.116 |
| 0.120 | 0.044 | 0.063 | 0.107 | 0.862 | 0.110 | |
| 0.152 | 0.048 | 0.050 | 0.099 | 0.855 | 0.103 | |
| 0.189 | 0.069 | 0.037 | 0.106 | 0.847 | 0.111 | |
| 0.210 | 0.085 | 0.034 | 0.119 | 0.835 | 0.125 | |
| HAP_IBD10 | 0.103 | 0.031 | 0.094 | 0.125 | 0.865 | 0.126 |
| 0.120 | 0.042 | 0.077 | 0.119 | 0.861 | 0.121 | |
| 0.152 | 0.051 | 0.055 | 0.106 | 0.865 | 0.110 | |
| 0.189 | 0.068 | 0.045 | 0.112 | 0.855 | 0.116 | |
| 0.210 | 0.084 | 0.045 | 0.129 | 0.847 | 0.132 |
Standard errors across replicates were calculated as the standard deviation of the estimated variances divided by 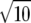 and ranged from 0.003 to 0.010 for the total genetic variance and from 0.002 to 0.008 for the residual variance.
and ranged from 0.003 to 0.010 for the total genetic variance and from 0.002 to 0.008 for the residual variance.
The sum of estimated QTL and polygenic variances.
The average simulated QTL and residual variances across replicates.
