Figure 3.
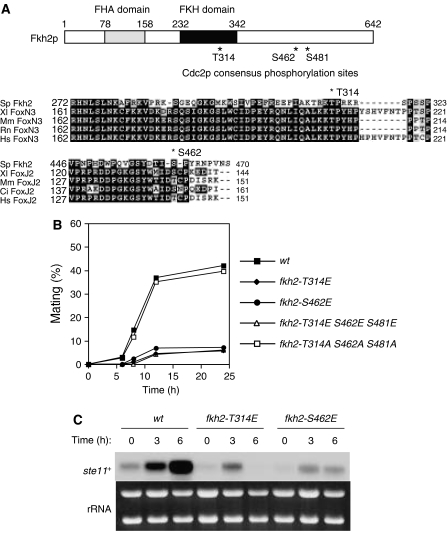
Phosphorylation of Fkh2p by Cdc2p negatively regulates mating. (A) A schematic representation of Fkh2p indicating consensus phosphorylation sites (T314, S462, S481) for Cdc2p as well as the FHA and FKH domains is shown in the upper panel. Multiple alignment of Fkh2, FoxN3, and FoxJ2 proteins of S. pombe (Sp), Xenopus laevis (Xl), Mus musculus (Mm), Rattus norvegicus (Rn), Homo sapiens (Hs), and Ciona intestinalis (Ci) is shown in the lower panels. Identical (shaded black) and similar (shaded gray) amino acids as well as two Cdc2p consensus phosphorylation sites (T314 and S462) of Fkh2p are indicated. Dashes represent gaps introduced to optimize alignment. (B) Cells expressing wild type (wt, HM5145) or T314E (fkh2-T314E, HM5910), S462E (fkh2-S462E, HM5911), T314E S462E S481E (fkh2-T314E S462E S481E, HM5827) or T314A S462A S481A (fkh2-T314A S462A S481A, HM5722) mutant forms of Fkh2p were treated and analyzed for mating efficiency as in Figure 1A. (C) Total RNA was extracted from cells treated as in (B) and was subjected to northern blot analysis of ste11+ mRNA.