Abstract
In previous studies we detected a frame shift mutation in the gene encoding the autoantigen La of a patient with systemic lupus erythematosus. The mutant La mRNA contains a premature termination codon. mRNAs that prematurely terminate translation should be eliminated by RNA quality control mechanisms. As we find Abs specific for the mutant La form in about 30% of sera from anti-La positive patients we expected that mutant La mRNAs circumvent RNA control and the expression of mutant La protein could become harmful. Indeed, realtime PCR, immunostaining, and immunoblotting data of mice transgenic for the mutant La form show that mutant La mRNAs are not repressed in these animals and are translated to mutant La protein. In addition to the mutant La protein, we detected a minor portion of native human La in the mutant La transgenic mice. Therefore, ribosomal frame shifting may allow the mutant La mRNA to escape from RNA control. Interestingly, expression of the mutant La mRNA results in a lupus like disease in the experimental mice. Consequently, escape of mutant La mRNA from RNA control can have two effects: It (i) results in the expression of an immunogenic (neo)epitope, and (ii) predisposes to autoimmunity.
Introduction
Eukaryotes have evolved a RNA quality control mechanism which allows cells to identify and degrade mRNAs containing nonsense mutations. This process is known as nonsense mediated decay of mRNA (NMD) or RNA surveillance (1,2). Nonsense mutations usually result in premature termination codons (PTC). The role of NMD is to recognize such mutant forms of mRNAs and to eliminate them as truncated mutant proteins could cause dominant negative effects on the normal cellular function of the respective native protein. Only in rare cases do mRNAs with nonsense mutations escape NMD. In all of these instances, the expression of the truncated proteins results in severe phenotypes. For example, the expression of mutant ROR2 mRNA leads to polydactyly, and expression of mutant forms of haemoglobins leads to β-thalaessemias (3–5).
The core NMD machinery is phylogenetically conserved from yeast to humans. Depletion of upf1 in mice – a factor which is assumed to be involved in the recognition of a PTC – leads to embryonic death at the implantation stage (6). This implies the fundamental and vital role of the NMD mechanism.
At the molecular level, NMD is not yet fully understood. According to recent models, so called exon junction complexes (EJCs) assemble at the splice junctions where they remain even after transport to the cytoplasm. During the first round of translation, these EJCs will be removed. If a mRNA contains a PTC, the ribosome does not pass all splice junctions and the EJCs downstream of the PTC will remain associated with the mutant mRNA. mRNAs containing EJCs after the initial round of translation will be degraded.
In previous studies we prepared an expression cDNA library constructed from peripheral blood mononuclear cell (PBMC) mRNA taken from an autoimmune patient (index patient) with systemic lupus erythematosus (SLE) (7–10). The cDNA library was screened with the patient’s own autoimmune serum which contained predominantly Abs to the autoantigen La/SS-B. During screening of the patient’s cDNA library we identified a mutant La cDNA. Genomic DNA analysis of serial blood drawings of the index patient shows that the mutation persists for more than a decade (Bachmann, unpublished). The mutation occurred in a region sensitive for mutations and can be found in more than 15 % of anti-La positive autoimmune patients (Semsei, personal communication).
Native La protein is thought to be involved in essential housekeeping functions. These functions include a role in transcription/termination of RNA synthesized by polymerase III, 3′-RNA processing, and nuclear RNA import and retention (11–17). Furthermore, this autoantigen may participate in stabilization of mRNAs and translation of viral and mitogen-stimulated mRNAs (18–29). E.g. La protein was shown to be involved in the cap-independent translation of the mRNAs encoding the X-linked inhibitor of apoptosis (XIAP) and the double minute gene product mdm2 which is a regulator of the stability of p53 (30–32). Thus, an impaired expression of La protein could even have a direct effect on the survival of autoreactive cells and could, thereby, contribute to the development of autoimmunity.
Due to the presence of the PTC in the mutant La mRNA, however, the mutant La mRNA should be eliminated by the NMD mechanism and should not be translated to protein. Consequently, it should not become a target of an immune response. However, here we report that the serum of this patient and about 30% of anti-La positive sera of SLE patients in general contain Abs to a mutant-La specific neoepitope suggesting that mutant La mRNAs were transcribed in these patients, circumvented NMD, were translated to protein and became a target of an immune response. In order to analyse how mutant La mRNAs escape NMD we established mice transgenic for the mutant form of La. We show, that mutant La mRNAs are not only repressed and translated to mutant La protein, but, unexpectedly, expression of mutant La favors the development of systemic autoimmunity.
Materials and Methods
Patient sera and mAbs
The serum samples from patients with SLE or primary Sjögren’s syndrome positive for IgG Abs to La protein were obtained from a Routine Autoantibody Testing Laboratory (Institute of Immunology, University of Dresden, Germany). In addition, we used serum samples from mothers of children with either cardiac or cutaneous manifestations of neonatal Lupus (ages 20–50 years) that were obtained from the Research Registry for neonatal lupus (33). The sera which were originally characterized as La positive by commercial routine ELISAs and immunoblotting were re-evaluated by ELISA and immunoblotting using recombinantly expressed human La protein (see also Proteins and immunoassays). Age- and sex-matched control sera from healthy individuals and anti-La negative sera from patients with SLE or scleroderma were acquired from the OMRF serum repository and the European Routine Autoantibody Testing Laboratory (Institute of Immunology, University of Dresden). The OMRF Institutional Review Board approved the use of all specimens in compliance with NIH guidelines. As mAbs we used the anti-La mAbs SW5, 4B6 and 3B9. SW5 is a well established anti-La mAb which recognizes the N-domain of human La protein but not mouse La protein (15). The anti-La mAb 4B6 was described previously and shown to be directed to the C-domain of human La (34). The anti-La mAb 3B9 was established previously in our lab and recognizes both human and mouse La protein (Bachmann, unpublished).
Mice
We housed all mice in specific pathogen free conditions, and the OMRF Institutional Animal Care and Use Committee approved all mouse-related studies. Mice transgenic for the wild type human La gene including its natural promoter have been described (39), and we used them as heterozygotes at the ≥F12 backcross generation. We genotyped these mice as previously described (35). Mutant La-transgenic mice were generated as follows. We initiated the creation of the mutant La transgene with a construct containing the native human La gene that we had also used to establish the mouse 3T3 cell line expressing native human La (8). First, we deleted an AvrII fragment from the native La genomic clone. The remaining construct contained a unique KpnI site in exon 3 and a unique BstEII site in exon 10 of La. We replaced the KpnI/BstEII fragment from this genomic native La construct with the respective mutant La cDNA fragment and then restored the AvrII fragment in the AvrII site. To prepare linearized DNA that was devoid of bacterially derived sequence for FVB/N fertilized oocyte injection, we released the mutant La gene from the vector with XhoI. We identified four positive FVB/N founder mice (one male and three females) by PCR analysis of tail DNA from a total of 81 animals and crossed them with FVB/N mice obtained from the Jackson Laboratory (Bar Harbor, ME). The male founder was used to establish the mouse strain M1. Two of the female mice did not transfer the transgene to their offspring. The remaining female founder produced a single positive F1 mouse. This mouse was used to establish the mouse strain M2. We checked the presence of the transgene by PCR using three primer pairs. The primer pairs were specific for regions either inside or at the 5′- or 3′-end of the human La gene (data not shown). We confirmed the presence of the transgene by Southern blotting (data not shown) and, ultimately, by analysis of protein expression.
Immunostaining and confocal microscopy
For immunohistochemistry tissue samples (approximately 0.25 cm2) were extracted from mice following CO2 euthanasia, fixed overnight in chilled 2% paraformaldehyde in PBS, then allowed to equilibrate in 30% sucrose at 4°C. Sucrose-infiltrated samples were cryopreserved in Tissue Freezing Medium (Triangle Biomedical Sciences, Inc., Durham, NC.) and 5 km serial sections were cut and placed on positively-charged slides. Slides were allowed to come to room temperature, then sections were treated 30 min with 0.3% hydrogen peroxide to block endogenous peroxidase activity. Sections were stained using the Histomouse™ SP DAB staining kit (Zymed Laboratories, Inc., South San Francisco, CA) according to manufacturers suggested guidelines. Briefly, sections were blocked for 40 minutes with BEAT™ blocking reagent then rinsed with water and PBS. Either PBS or a 1 to 50 dilution of the anti-human La mAb SW5 was added to the sections and incubated at room temperature in a humidified chamber for 1h. After rinsing with PBS, sections were incubated for 10 min with biotinylated enzyme conjugate, developed with the substrate-chromagen mixture for 5 min, and rinsed with distilled water. Slides were counterstained with hematoxylin for 1 min, rinsed thoroughly in tap water followed by 30s in PBS then distilled water. Sections were dehydrated through a graded alcohol series and three changes of xylene and coverslipped. Slides were evaluated on the Zeiss Axioplan 2i light microscope (Carl Zeiss Microimaging Inc., Thornwood, NY) at 63X and 100X magnifications and photodocumented.
For detection of immune complexes in kidney sections kidneys were fixed in chilled 2% paraformaldehyde in PBS and equilibrated in 30% sucrose in PBS prior to cryopreservation. We then blocked 5 μm sections with 5% powdered milk in PBS and subsequently stained with optimized dilutions of primary conjugates of Alexa Fluor 546® rabbit anti-mouse IgG (H+L) (Molecular Probes, Eugene, OR) and goat anti-mouse complement C3-FITC (ICN-Cappel, Aurora, OH) in PBS. We rinsed the samples twice with PBS containing 0.1% Triton X-100, twice with PBS alone, then incubated with an optimized dilution of 4′,6-Diamidino-2-phenylindole (DAPI) (Sigma, St. Louis, MO), and rinsed twice in PBS. We then visualized fluorescence using a Zeiss LSM 510 Meta Confocal Microscope (Carl Zeiss International, Thornwood, NY).
Proteins and immunoassays
For immunoassays, we expressed the recombinant 6xhistidine-tagged human La from a pET3d vector. The C-domain of La protein and the N-domain of La either lacking or containing the neoepitope sequence were cloned in a pET28a vector and also expressed as 6xhis-tagged fusion proteins (34). Bovine Ro was obtained from Kevin Clark (ImmunoVision, Springdale, AR), BSA was purchased from Sigma (St. Louis, MO). The University of Oklahoma Health Science Center Molecular Biology Core Facility prepared synthetic peptides as eight-branched muntiantigenic peptides (MAPs) (neoepitope: AKKMKKENKIKWKLN) and purified them by HPLC. Screening of human and mouse sera for Abs to extractable nuclear autoantigens (Sm, RNP, Ro, La, Scl-70, Jo-1) was performed using commercial ELISA kits such as the RELISA multiparameter screening test (Immuno concepts, Sacramento, CA). For further evaluation, we coated ELISA plates with 50 kl of the respective purified native or recombinant antigen including La, Ro, Sm, RNP, Scl-70, Jo-1 (5 μg/ml in 0.15 M Na2CO3, 0.35 M NaHCO3 carbonate buffer, pH 9.6) or synthetic peptides (1 μg/ml in carbonate buffer) for 2 h at room temperature or overnight at 4°C, washed them briefly with 0.1% Tween-20 in PBS, then blocked them with 0.1% gelatin in PBS, pH 7.4, overnight at 4°C. After washing with 0.1% Tween-20 in PBS, we added 50 kl sera diluted (1/1000) in PBS and incubated the plates for 2 h at room temperature. We then washed the wells with 0.1% Tween-20 in PBS, incubated the wells with alkaline phosphatase-labeled anti-human (Jackson Immunoresearch, West Grove, PA) or anti-mouse IgG (Sigma, St. Louis, MO), and detected bound Abs by addition of the substrate paranitrophenylphosphate (pNPP). As positive controls we analyzed monoclonal (if available) and patient Abs to La, NeoLa, Ro, RNP and Sm in parallel on each ELISA plate. The absorbance was measured at A410.
Serum samples for Abs to dsDNA were assayed with a commercial ELISA Kit from Alpha Diagnostic International (San Antonio, Texas, Cat. No. 5100). This Kit was specifically developed for detecting anti-dsDNA Abs in mouse serum. According to the manufacturer, serum samples known to be positive for anti-extractable nuclear antibodies, anti-ssDNA, anti-rheumatoid factor, anti-toxoplasma gondii IgG, IgM and anti-cytomegalovirus IgG are negative.
Immunoprecipitation and SDS-PAGE/immunoblotting
For immunoprecipitation of total extracts we used the immunoprecipitation system from NatuTec (Frankfurt, Germany). Immunoprecipitated proteins were subjected to SDS/PAGE immunoblotting. The NatuTec system uses a secondary anti-mouse Ab which is directed to a conformational epitope that is destroyed under reducing conditions. Consequently, heating of the immunoprecipitated material prior to electrophoresis under reducing conditions destroys the ability to detect coprecipitated mouse immunoglobulins. Blots were qualitatively and quantitatively evaluated using the Enhanced Chemiluminescence (ECL) system (Amersham, Freiburg, Germany) and the CHEMI DOC XRS system (Bio-Rad, Munich, Germany).
Quantitative reverse transcription-PCR
The expression of human exon 1, exon1′, 1″, mouse exon 1a,b,c La mRNA, and actin mRNA was analyzed by a light cycler (LC)-based PCR assay. The relative mRNA quantity was determined applying a realtime PCR protocol based on SYBR Green I detection (LC - FastStart DNA Master SYBR Green I; Roche Diagnostics, Mannheim, Germany). Serial dilutions of plasmid DNA containing the respective specific human or mouse La mRNA or actin fragment over seven log scales (102–108 molecules per capillary) were used as internal template standards (calculation via a light cycler (LC) quantification software version 3.5; Roche Diagnostics, Mannheim, Germany). Each determination was carried out twice for each cDNA sample as independent PCR runs. The mean values for the La mRNA forms were normalized to the actin control. The assays were first established on normalized commercial human (I, II, fetal, and blood fractions) and mouse cDNA panels (I and II; all from BD Biosciences Clontech, Palo Alto, CA). After subcloning of the PCR products in pGEM-Teasy the specificity of the PCR reactions was determined by sequencing. Finally, mRNAs were isolated from shock frozen tissues of native human La, mutant human La and non-transgenic mice. mRNA isolation and cDNA synthesis was performed as described previously (15). As forward primer we used for amplification of human exon 1 La (5′-GGAGTCGTTGCTGTTGCTGTTTGTG), human exon 1′La (5′-GGGGTAAACGCCGGAGGGTTC), mouse exon 1a,b La (5′-GGAAGTCCAGGCGCTTCTGTCG), mouse exon 1c La (5′-GGGAAACCTGTAAGGT TAGGAATTC), and actin (5′-GCCGTCTTCCCCTCCATCGTG) mRNA. As reverse primer we used for amplification of human exon 1 and 1′ La (5′-CACTGATTTCCATGAGTTCTGCCTTGG), mouse exon 1a,b/c La (5′-GGGAGTGGTTGCTTGGTGATCTT), and actin (5′-GGAGCCACACGCAGCTCATTGTAGA) mRNA. The PCR protocol consisted of a pre-denaturation step (10 min at 95°C) and 40 amplification cycles (15s at 95°C for all samples; for human exon 1 La: 5s at 74°C; for human exon 1′ La and actin mRNA: 5s at 70°C; for mouse exon 1a,b La mRNA: 5s at 68°C; for mouse exon 1c La mRNA: 5s at 64°C; 14s at 72°C for all samples).
Results
Mammalian La proteins consist of two domains (Fig. 1a). The N-domain binds to small RNAs. The C-domain contains the nuclear location signal (NLS). In human La the peptide which connects the two domains is encoded by exon 7. Within exon 7, the La sequence contains an oligo(A)8-region (Fig. 1b, nLa, nts 1051–1059) which encodes the aa 190–192 (Fig. 1b, nLa, aa KKN). The previously isolated mutant La cDNA has a deletion of an (A)-residue in the oligo(A)8- region (Fig. 1b, muLa, arrow). The frame shift mutation results in a PTC at nts 1091–1093. Downstream of the deletion, the aa sequence of the truncated mutant form of La protein (muLa, Fig. 1b, aa 192–204) differs from the native La sequence (nLa). As schematically summarized in Fig. 1 (c1, c2), La protein is encoded by twelve exons. Consequently, eleven introns must be removed during splicing from the primary La gene transcript. As four exon junctions follow downstream of the PTC the mutant La mRNA should be recognized by NMD.
FIGURE 1.
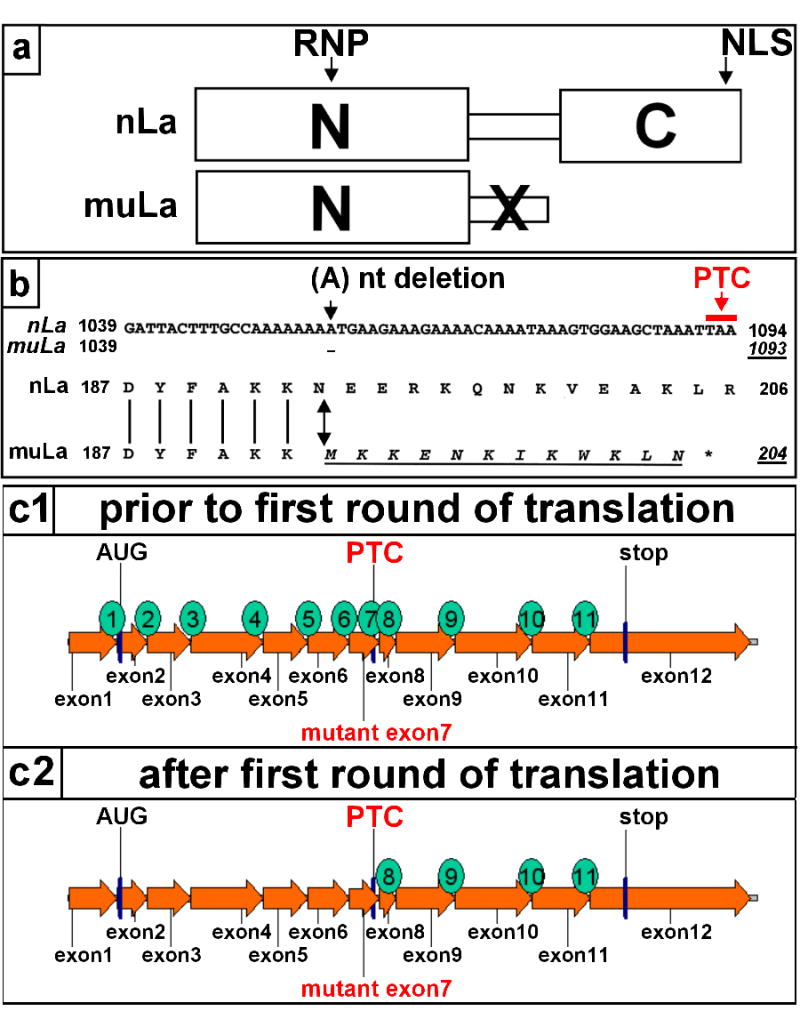
Native (nLa) and mutant human La (muLa) protein (a,b). (a) nLa consists of two domains. The N-domain contains a RNP consensus motif. The C-domain contains a nuclear localization signal (NLS). muLa contains a frame shift mutation (X). The resulting mutant protein reading frame is truncated and lacks the NLS. (b) nLa) contains eight (A) residues at position 1051 to 1058. The deletion of one (A)-residue (muLa, arrow) results in a PTC (nt 1092–1094). Thus, the mutant La mRNA encodes a mutant reading frame (muLa) which differs from the native La reading frame (nLa) downstream of the frame shift mutation (double headed arrow). The mutant aa sequence is underlined. The “*” denotes the location of the PTC (c1, c2) According to recent models, mRNAs containing a PTC are recognized and eliminated by NMD as EJCs will remain associated with the mRNA after the first round of translation. As schematically shown in c1, the native human La mRNA consists of 12 exons. Consequently, 11 splice junctions will be formed during splicing. Four of these junctions locate downstream of the PTC (c2). Consequently, these four EJCs should remain associated with the mutant La mRNA after the first round of translation and, thus, the mutant La mRNA should be recognized and destroyed by NMD.
Detection of Abs to a neoepitope in mutant La in sera of anti-La positive autoimmune patients
As mentioned above, downstream of the mutation the mutant La aa sequence MKKENKIKWKLN (Fig. 1b, muLa, aa 193–204) differs from the native La aa sequence EERKQNKVEAK (Fig. 1b, nLa, aa 193–203). Full length La protein, the native 29 kD form of the N-domain of La (Fig. 2a: LaN; aa 1 to 203), the mutant form of the N-domain of La (Fig. 2a: LaN-Neo; aa 1 to 204), and the native 25 kD C-domain of La (Fig. 2a, LaC; aa 204–408) were recombinantly expressed and blotted against serum samples from either healthy donors (50 samples) or anti-La negative (25 samples) or anti-La positive autoimmune patients (100 samples). The respective anti-La Abs were visualized with anti-human IgG specific secondary Abs which did not cross-react with human IgM antibodies. About 15% of the anti-La positive sera reacted with LaN-Neo but not with LaN suggesting a specific reactivity to the La neoepitope in these serum samples. One example is shown in Figure 2a (lane 4). In addition to the anti-LaN-Neo reactivity, some sera reacted with the C-domain of La (e.g. in Fig. 2a, lane 9). Most of interest are the results obtained for the serum samples shown in Fig. 2a (lanes 4 to 6). These serum samples represent serial drawings from the same patient. The second sample (Fig. 2a, lane 5) was taken six month, the third sample (Fig. 2a, lane 6) one year after the first drawing (Fig. 2a, lane 4). These immunoblotting data show that the anti-mutant La response preceded the anti-La response in this patient.
FIGURE 2.
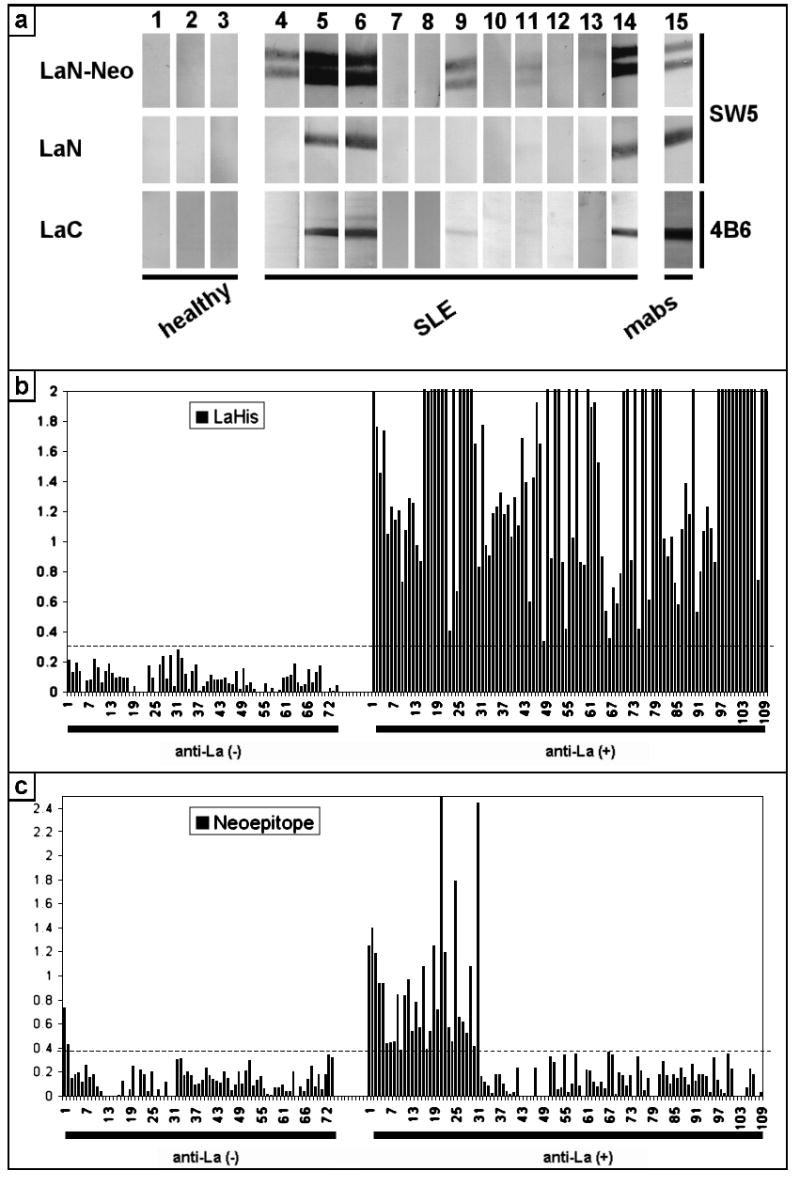
Anti-NeoLa Abs in autoimmune patients. (a) The native N-domain (LaN), the mutant N-domain (LaN-Neo), and the native C-domain (LaC) of La protein and full length La protein was recombinantly expressed and purified using Nickel-affinity chromatography. The purified fragments were dialysed against PBS. During dialysis various amounts of the La fragments undergo proteolysis resulting in some cases in truncated proteins. All La fragments were blotted against serum samples of either healthy donors (e.g. lanes 1 to 3) or autoimmune patients (e.g. lanes 4 to 14). In addition, the recombinant proteins were blotted against anti-La mAbs directed to either the N-domain (lane 15, SW5) or the C-domain (lane 15, 4B6) of La protein. (b) 74 anti-La negative and 109 anti-La positive sera were analyzed by ELISA using recombinant human La protein as substrate. (c) The same sera as in (b) were analyzed for anti-NeoLa peptide antibodies.
It is obvious including from the results shown in Fig. 2a (lanes 5,6 and 14), that the immunoblotting technique does not allow us to identify the presence of Abs specific for LaN-Neo in serum samples which react with the native N-domain of the La antigen. Therefore, we expected to underestimate the percentage of sera specific for LaN-Neo with the immunoblotting technique. In order to overcome this disadvantage of the immunoblotting assay, we developed a peptide ELISA specific for mutant La. For this purpose we designed a series of overlapping synthetic peptides including the NeoLa peptide (AKKMKKENKIKWKLN, the mutant La portion is underlined) and the corresponding native La peptide NatLa (FAKKEERKQNKVEAK). We analyzed 109 anti-La positive sera (Fig. 2b, LaHis, anti-La(+)) at dilutions of 1 to 1000 for Abs to the NeoLa (Fig. 2c, Neoepitope, anti-La(+)) and NatLa peptide (data not shown). Thirty-one sera reacted with the NeoLa peptide at levels greater than 4 standard deviations above the mean. In contrast, only two (n=74) of control negative sera were positive. Our negative control group consisted of anti-La negative SLE patients (n=52, 20 sera were anti-Ro only), patients with scleroderma (n=7) and healthy blood donors (n=15). None of the sera reacted with the NatLa peptide although it contained the three aa AKK that were also part of the NeoLa peptide. In order to confirm the specificity of the anti-NeoLa-peptide reactivity we adsorbed anti-NeoLa positive sera with the recombinantly expressed mutant La protein which completely blocked the anti-NeoLa reactivity (data not shown). In contrast, the recombinantly expressed N-domain of La lacking the NeoLa sequence was not able to block the anti-NeoLa peptide reactivity. Moreover, NeoLa positive sera were negative when analysed on peptides representing the scrambled NeoLa amino acid sequence (data not shown).
Taken together, our data show that (i) a mutant La response can precede an anti-La response, (ii) Abs to mutant La can be detected in about 30% of anti-La positive autoimmune patients, and (iii) mutant La can become a target of an immune response.
Development of two independent mouse lines transgenic for a mutant human La gene
The expression of mutant La protein requires that mutant La mRNA is capable of escaping NMD. In order to provide experimental evidence for an escape of mutant La mRNA from NMD we established mice transgenic for mutant human La. The genomic construct encoding mutant human La was cloned as schematically summarized in Fig. 3 (a) and described in detail in Materials and Methods. In summary, we replaced a KpnI/BsTEII fragment in a reconstructed human La gene clone with the respective KpnI/BsTEII fragment from a mutant La cDNA clone. The mutant exon7 was part of this KpnI/BsTEII fragment.
FIGURE 3.
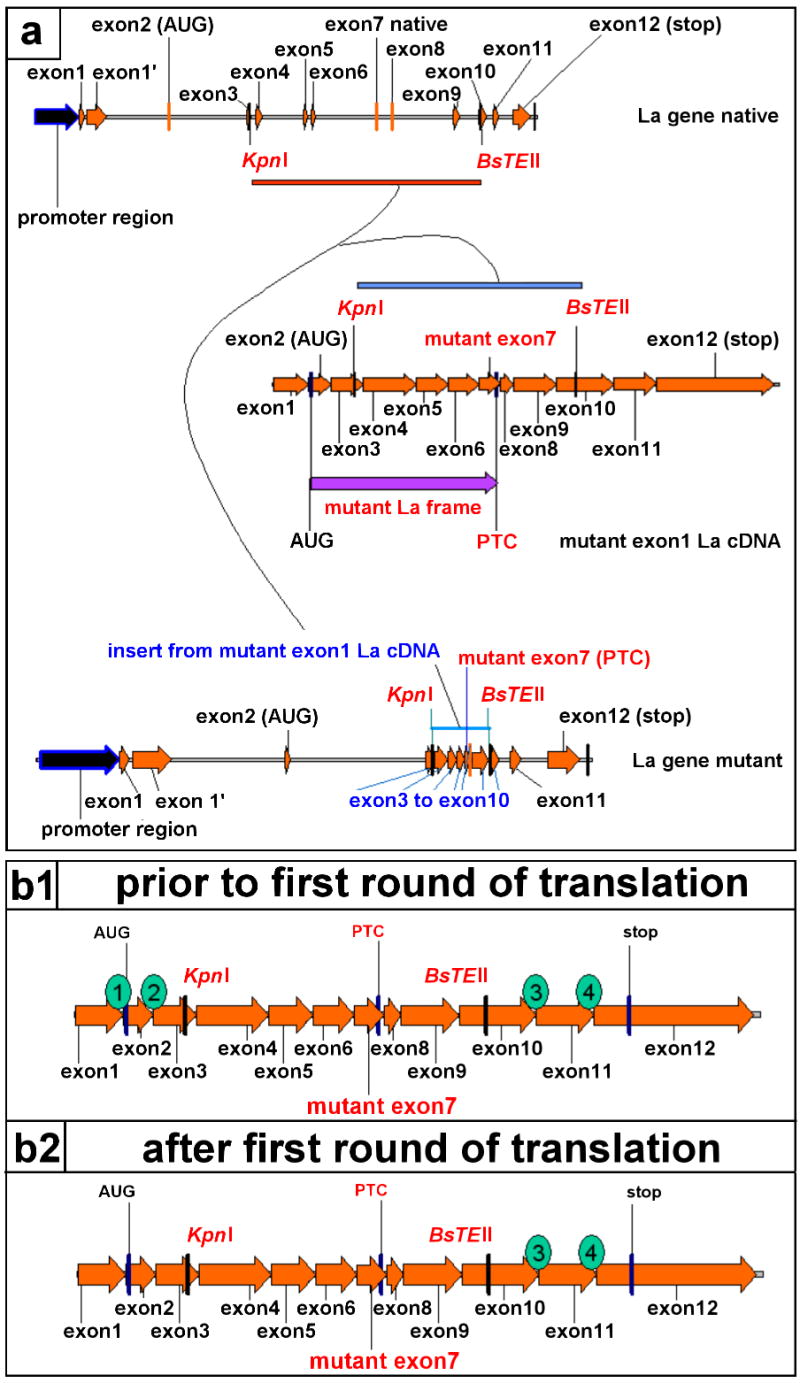
Features of the mutant La transgene. (a) The cloning of the mutant La transgene (La gene mutant) started from a native human La transgene construct (La gene native). We replaced the genomic region from exons 2 to 10 of the human native La transgene with the respective sequence of a mutant La cDNA (mutant exon 1La cDNA). For this purpose we isolated the KpnI/BsTEII fragment from the mutant exon 1 La cDNA construct and inserted it in the respective sites of the native La transgene. (b1, b2) Thereby, we conserved the introns upstream and downstream of the cDNA inserts. A mutant La mRNA transcribed and processed from this construct contains four splice junctions (1 to 4). Two splice junctions locate upstream and two downstream of the PTC. During the first round of translation the scanning ribosome should not pass the exon junctions 3 and 4. Consequently, two EJCs should persist downstream of the PTC and the mutant La mRNA should be recognized and eliminated by NMD.
It is obvious that the cloning procedure altered the number of splice sites in the resulting mutant La gene construct. Nonetheless, two splice sites were conserved downstream of the PTC and, thus, the encoded mRNA should still be recognized by NMD as schematically shown in Fig. 3b.
Two independent transgenic mouse lines (M1 and M2) were established from the mutant La gene construct (Fig. 3a). The integration of the transgene was detected by PCR and Southern blotting (data not shown).
Analysis of expression of mutant La at the RNA level in mutant La transgenic mice
Realtime PCR assays were established allowing us to measure the copy numbers of human and mouse La mRNAs. In previous studies we had shown that three La mRNA isoforms can be transcribed from the human La gene which we termed exon 1, exon 1′ and exon 1″ La mRNAs. Comparable studies were not available for the mouse La gene. Therefore, we analyzed the expression of the mouse La gene (Bachmann, unpublished). From these studies we know that three mouse La mRNA isoforms exist. We termed these isoforms exon 1 a, b and c. Like all the human La mRNA isoforms, all the murine La mRNAs are also functional. In spite of their different 5′-starts, all La mRNAs encode the same human or mouse La protein, as the initiation codon locates in the exon 2.
Quantitative PCR was performed for RNAs isolated from tissues of either mutant La transgenic (Fig. 4a, M1, M2; Fig. 4b, muLa), or non-transgenic (Fig. 4b, non-tg), or native La transgenic mice (Fig. 4b, nLa). In addition, we estimated the copy number of La mRNAs for commercially available multi-tissue–cDNA panels from mouse (Fig. 4b, mMTC) and human (Fig. 4b, hMTC) tissues. We analyzed a total of eight mutant La transgenic mice, four native La transgenic mice, and four non-transgenic FVB mice. RNAs were obtained from brain, liver, spleen, gut, and kidney. In addition, we analyzed the RNAs from thymus tissue when available. Results for liver and spleen tissues are shown in Fig. 4(a,b). Fig. 4(a) shows the data for individual animals. Fig. 4(b) gives the mean values of all analyzed animals. Our data shows: (i) the mouse La mRNAs are expressed at similar levels in non-transgenic and transgenic animals, (ii) the human La mRNAs in the transgenic mice are expressed in addition to the endogenous mouse La mRNAs and, (iii) the expression level of the respective La transgene is comparable to the endogenous mouse La mRNA level. Consequently, (i) the expression of the respective transgene does not alter the expression of the mouse La gene and (ii), the mutant La transgene is expressed at similar levels as the native La transgene. Thus, the mutant La mRNA escapes NMD.
FIGURE 4.
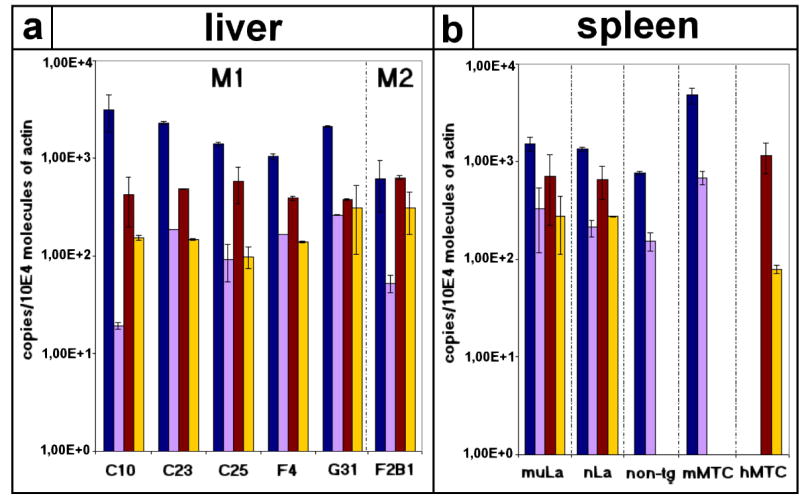
Quantitative analysis of expression of mutant La mRNA in mutant La transgenic mice. Quantitative PCR was performed for RNAs isolated from liver (a) or spleen (b) tissues of either mutant La transgenic (a, M1, M2; b, muLa), or non-transgenic (b, non-tg), or native La transgenic mice (b, nLa). In addition, we estimated the copy number of La mRNAs for commercial multi-tissue–cDNA panels from mouse (b, mMTC) and human (b, hMTC). We estimated the copy numbers for mouse exon 1a and b La (blue bars), mouse exon 1c La (purple bars), human exon 1 La (brown bars), and human exon 1′,1″ La (yellow bars) mRNAs. The copy numbers were normalized to actin. (a) shows the data for liver tissues of six individual animals. The estimations were performed at least as duplicates. The error bars show the deviation of the values. (b) shows the data for spleen tissues. The mean values and the standard deviations of all analzyed animals are given. Similar results were obtained for other tissues.
Analysis of expression of mutant La in extracts from tissues of mutant La transgenic mice
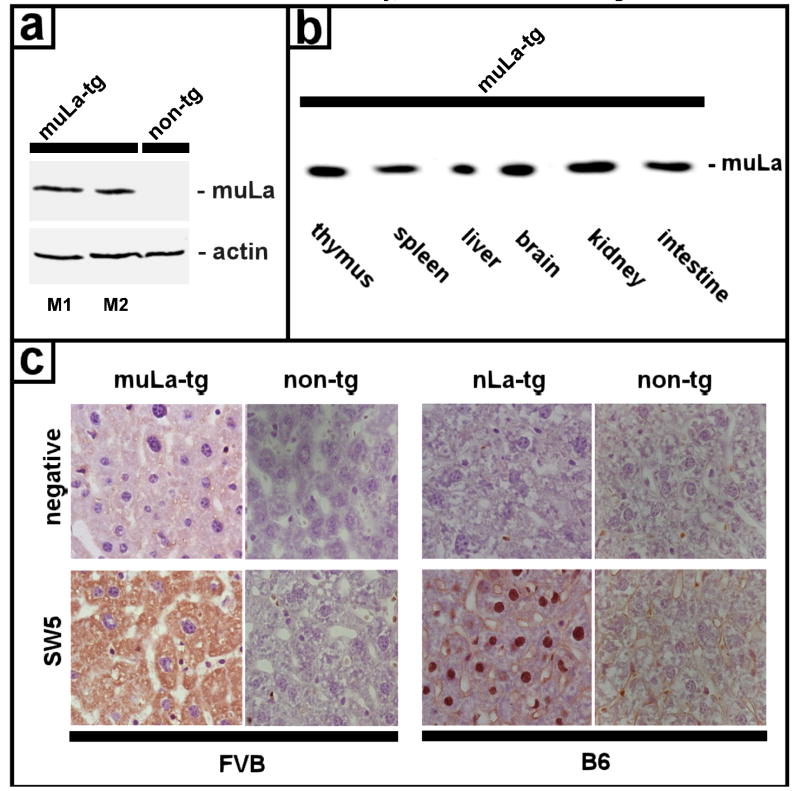
Using the anti-human La specific mAb SW5 we detected the truncated human La protein in total tissue extracts of both mutant La-transgenic mouse lines M1 and M2 (Fig. 5a, muLa-tg, M1 and M2). In order to compare the expression levels, extracts of mutant La transgenic mice (Fig. 5a, muLa-tg) and non-transgenic mice (Fig. 5a, non-tg) were normalized with anti-actin antibodies. For some total tissue extracts, the detection of the mutant La transgene was difficult because of a high content of mouse immunoglobulins in these extracts which cross-reacted with the secondary Abs (data not shown). In order to improve the quality of the immunoblots and to allow a quantitative comparison of the mutant La protein expression in different tissues, we immunoprecipitated the extracts with the anti-human La specific mAb SW5 and detected the immunoprecipitated antigen with a secondary Ab which does not bind to reduced immunoglobulins (see Materials and Methods). As shown in Fig. 5(b) the mutant La protein was equally expressed in all organs tested including in thymus, spleen, liver, brain, kidney, and intestine (Fig. 5b, muLa).
FIGURE 5.
Expression of mutant La protein in transgenic mice. (a) Extracts from mutant La transgenic mice (muLa-tg; M1,M2) and non-transgenic mice (non-tg) were prepared from spleen tissues and analyzed by SDS-PAGE/immunoblotting with the anti-La mAb SW5 which recognizes the N-domain of human La. The extracts were normalized with anti-actin antibodies. (b) Extracts from different tissues of mutant La transgenic mice (muLa-tg) were prepared from thymus, spleen, liver, brain, kidney and intestine, immunoprecipitated with the anti-La mAb SW5 and analyzed by SDS-PAGE/immunoblotting with the anti-La mAb SW5 which recognizes the N-domain of human La and therefore also reacts with mutant human La (muLa). (c) Tissues from mice transgenic for mutant La (muLa-tg) or native La (nLa-tg) or from non-transgenic mice (non-tg) were stained with the human anti-La specific mAb SW5 (see Materials and Methods). Due to the lack of the NLS, mutant La (muLa-tg, SW5) is in the cytoplasm while native La (nLa-tg, SW5) is in the nucleus. SW5 does not stain tissues from non-transgenic mice irrespective of the background (FVB/N or B6). The negative controls (negative) were also not stained.
Intracellular localization of mutant La in tissues of mutant La transgenic mice
When tissues were stained with the anti-human La specific anti-La mAb SW5, the mutant La protein which lacks the NLS was found in the cytoplasm (Fig. 5c, muLa-tg) while the native La transgene was found in the nucleus (Fig. 5c, nLa-tg).
Detection of native human La in extracts from tissues of mutant La transgenic mice
Extracts from tissues of mutant La transgenic (Fig. 6, lanes a to d), native La transgenic (Fig. 6, lanes e,g,i), and non-transgenic (Fig. 6, lanes f,h,j) mice were precipitated with an anti-human La serum. The precipitated La peptides were analyzed with the anti-La mAbs 3B9 (Fig. 6, lanes a,e,f), SW5 (Fig. 6, lanes b,g,h), and 4B6 (Fig. 6, lane c,i,j). The anti-La mAb 3B9, which recognizes the N-domain of both human and mouse La protein, reacted with the endogenous mouse La protein in all analyzed transgenic and non-transgenic extract samples (Fig. 6, lanes a,e,f). In addition, it reacted with the mutant human La protein in the extract of the mutant La transgenic mouse (Fig. 6, lane a) and the native human La protein in the extract of the native La transgenic mouse (Fig. 6, lane e). The human anti-La specific mAb SW5, which is directed to the N-domain of human La, also reacted with the mutant human La protein (Fig. 6, lane b) and the native human La protein (Fig. 6, lane g) but failed to react with the endogenous mouse La protein (e.g. Fig. 6, lane h). The anti-human La specific anti-La mAb 4B6, which is directed to the C-domain of human La, also reacted with the native human La protein in the extract of the native La transgenic mouse (Fig. 6, lane i). However, it failed to react with the mutant human La protein (Fig. 6, lane c) and the endogenous mouse La protein (e.g. Fig. 6, lane j). Unexpectedly, all three monoclonal anti-La Abs reacted with an additional protein in the extract of the mutant La transgenic mice (Fig. 6, lanes a to c). This protein, which has the same molecular weight as full length human La protein, is absent in the extract of the non-transgenic mouse (Fig. 6, lanes f,h,j). Taken together, the protein (i) has the same molecular weight as human La protein, (ii) is coprecipitated by the anti-La serum, and (iii) is recognized by three anti-La mAbs. From previous studies we know that the three anti-La mAbs recognize independent epitopes. The two epitopes recognized by the anti-La mAbs 3B9 and SW5 are upstream of the frame shift mutation. The epitope recognized by the anti-La mAb 4B6 is downstream of the frame shift mutation. Therefore, we conclude that this protein represents full length human La protein. Full length human La protein can only be translated from the mutant La mRNA if ribosomal frame shifting occurs upstream of the PTC.
FIGURE 6.
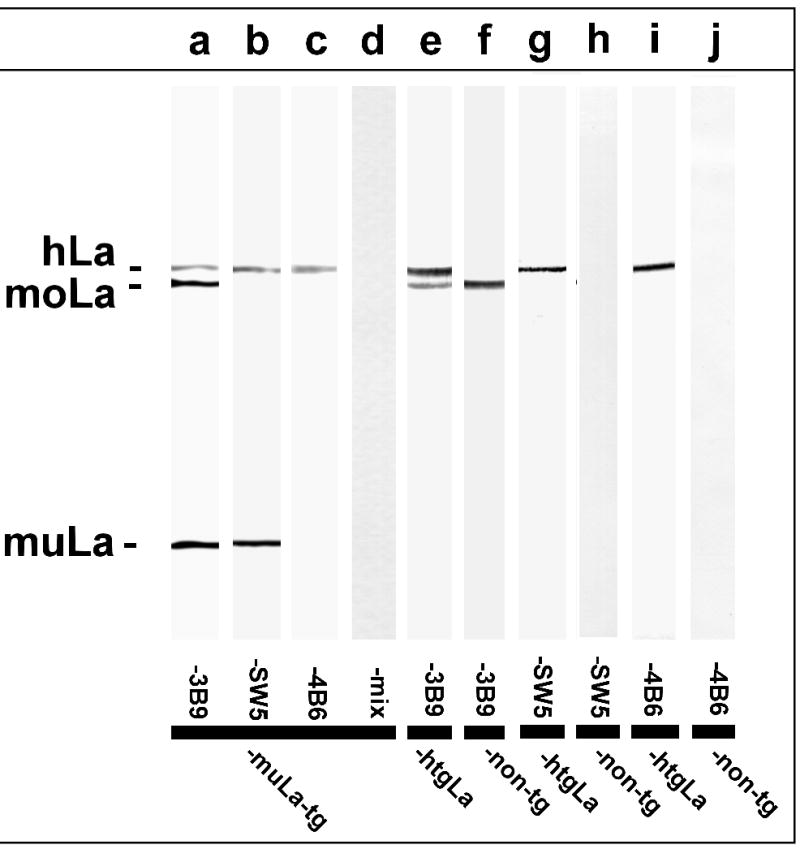
Expression of full length human La in mutant La transgenic mice. Total extracts from liver tissue of mice either transgenic for mutant La (lanes a to d), or native La (lanes e,g,i), or non-transgenic (lanes f,h,j) were immunoprecipitated with an anti-La serum and blotted against the anti-La mAbs 3B9 (lanes a,e,f), SW5 (lanes b,g,h), and 4B6 (lanes c,i,j). The anti-La mAb 3B9 recognizes human and mouse La protein. In contrast, the anti-La mAbs SW5 and 4B6 do not cross-react with mouse La protein. The epitopes recognized by SW5 and 3B9 locate in the N-domain of La while the epitope recognized by 4B6 locates in the C-domain. A mixture of unrelated mAbs did not react with any immunoprecipitated protein (lane d). hLa, human La; moLa, mouse La; muLa, mutant La.
Systemic autoimmunity in mice transgenic for mutant human La
As mentioned above, during the immunoblotting experiments we detected high levels of immunoglobulins in extracts from tissues of older mutant La transgenic mice, especially in kidney tissue extracts. Similar results were not seen for age-matched FVB/N controls (data not shown). Clinical SLE is characterized by the presence of anti-nuclear autoantibodies (ANA) and immune complex deposits in tissues.
Immune complex nephritis in mice transgenic for mutant human La
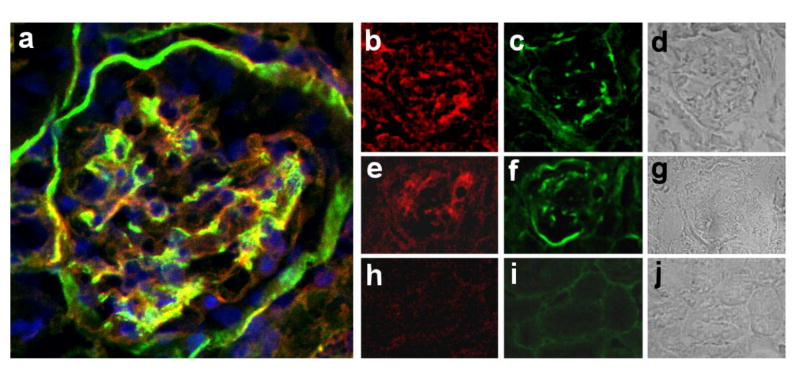
Therefore, we checked mutant La transgenic mice for proteinuria and analyzed kidney tissues of mutant La transgenic mice for Ab deposits. Six out of ten animals showed mild to moderate proteinuria. Two animals had severe proteinuria (+++). Tissues from these animals were further analyzed. Their histology data are shown in Fig. 7. We detected glomeruli which were positively stained with Abs to Ig and complement (Fig. 7a to g). Similar deposits were not seen in age-matched FVB/N controls (Fig. 7h to j).
FIGURE 7.
Immune complex deposition in kidneys of mutant La-transgenic mice. We stained kidney sections from mice transgenic for mutant La (a to g) and FVB/N control animals (h to j) with anti-Ig Ab (red), anti-complement Ab (green), and DAPI (blue). (a) Three colour overlay of a nephron of a mouse transgenic for mutant La. (b to g) Glomeruli from mice transgenic for mutant La. (h to j) FVB/N controls.
Spontaneous development of autoantibodies
The histology data suggested the development of autoantibodies in mutant La transgenic mice. We analyzed 58 serum samples taken from 42 mutant La transgenic mice (Fig. 8a,b) and 15 serum samples of age matched non-transgenic FVB/N control mice for Abs to NeoLa, La, Ro, Sm, RNP, and dsDNA. Forty out of 42 serum samples, taken from 38 mutant La transgenic animals which were older than five months and younger than eight months, contained autoantibodies to these nuclear antigens. All animals (n=18) older than eight months, including the two mice that were negative at the age of 5 months, contained autoantibodies. Only few animals younger than four months (n=16) had autoantibodies.
FIGURE 8.
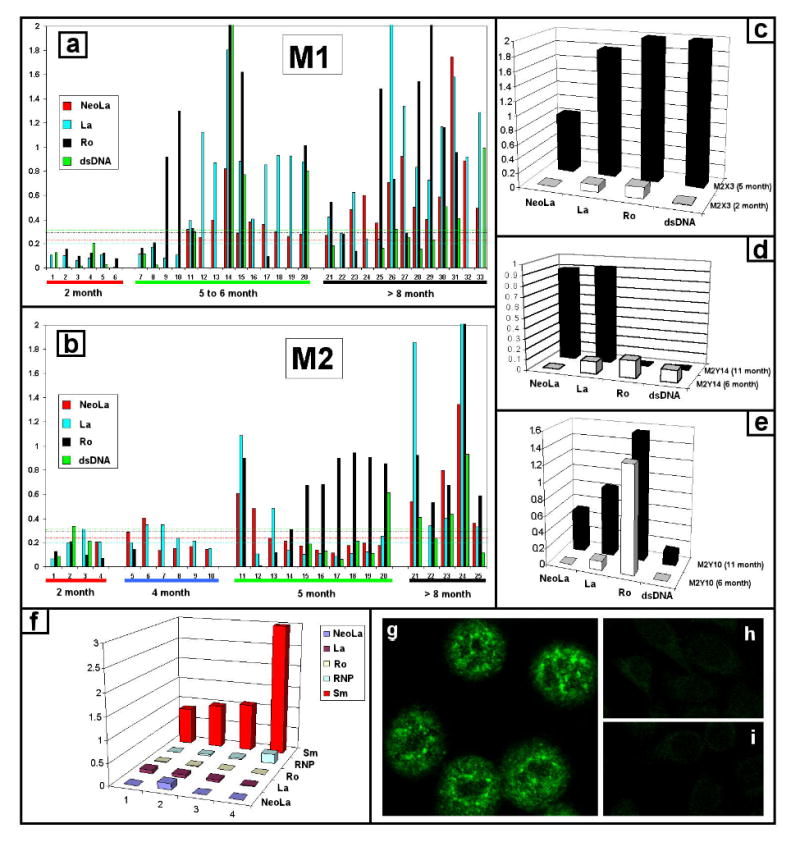
Autoantibodies in mice transgenic for mutant La. (a,b) Development of autoantibodies in mice transgenic for mutant La. We analyzed sera from the mutant La-transgenic mouse lines M1 and M2 for Abs to neoLa, La, Ro and dsDNA. The dashed lines represent cutoffs for positive reaction based on mean plus four standard deviations of 15 control FVB/N anti-sera. Nine of the control mice were five to six months of age. Six mice were older than seven months. (c to f) Ab pattern of individual mice. The autoantibody pattern differs between individual mice. Some mice develop Abs to NeoLa, La, Ro and dsDNA (e.g. c), others Abs to NeoLa and La but not to Ro and dsDNA (e.g. d) or to NeoLa, La and Ro but not to dsDNA (e.g. e). In some mice the response started with anti-Ro Abs (e.g. e) in others with anti-Sm Abs (f). The serum samples (1 to 4) in f were taken at the age of 4 to 8 months. (g) One of the analyzed mice was positive for anti-nuclear Abs in immunofluorescence but negative in ELISA for Sm, RNP, Ro, La, dsDNA. (h) FVB/N negative control. (i) secondary Ab control.
In general, the autoantibody response varied and development of autoantibodies did not depend on homozygosity of the mutant La transgene. In detail, eight mice developed Abs to NeoLa, La, Ro and dsDNA (Fig. 8c, e.g. M2X3). Six mice developed Abs to NeoLa and La but not to Ro or dsDNA (Fig. 8d, e.g. M2Y14). In eight samples we detected anti-Ro60 Abs as the initial specificity (Fig. 8e, e.g. M2Y10). In one animal the Ro-only response did not change over several months, while in the others the anti-Ro response spread to NeoLa and La (Fig. 8e, M2Y10). One mouse developed only anti-Sm Abs (Fig. 8f). In another mouse (Fig. 8g) we could detect anti-nuclear Abs only by epifluorescence microscopy. None of the mice developed Abs to Scl70. The threshold for positivity was determined as the mean plus four SDs of the fifteen non-transgenic FVB/Ns. As positive controls for anti-dsDNA Abs we included sera of age-matched Fc-gamma receptor knockout mice (data not shown).
In none of the control mice which were older than five months and younger than eight months did we detect autoantibodies to the nuclear antigens Ro, La, Sm and dsDNA. One of six non-transgenic FVB/-N mice older than eight month developed anti-nuclear antibodies. However, these Abs were IgM-type Abs and were not specific for the lupus antigens La, Ro, Sm, RNP or dsDNA.
Finally, we wanted to learn whether or not the overexpression of native La can also result in an autoimmune phenotype. Therefore, we collected the sera of 25 mice which were transgenic for the native La antigen. All the sera were negative for anti-nuclear Abs including for Abs to La, Ro, Sm, RNP, Scl-70, and Jo-1 (data not shown). 12 of these sera were from animals older than eight month, six sera were from mice between 5 and 6 month, and seven sera were from mice of 2–3 month of age. Taken together, these data support our interpretation that the autoimmune phenotype of the mutant La transgenic mice is the result of the transgenic expression of the mutant La form but not an overexpression of La.
Discussion
In 1994 we prepared a cDNA library from tissue of an autoimmune patient with SLE and Sjögren’s syndrome. This cDNA library was screened with the autologous serum of the patient (7). Thereby we isolated a mutant form of a La cDNA (7–10). From analysis of serial blood samples, we know that the mutation persists in the patient for more than a decade. Thus, we suspect that the mutation exists in a memory or stem cell of the patient. However, for obvious reasons we do not know if the mutation already occurred during embryonic development and, thus, before development of tolerance or was acquired after the development of tolerance.
Since our development of the autologous cDNA expression library approach to detect self antigens, Sahin et al. (36) and many others have extended this technique to sera and tissues from tumor patients and termed it as serological identification of antigens by recombinant expression cloning (SEREX) (36). And, the list of autoantigens that are detected by the SEREX technique is still growing (www2.licr.org/CancerImmunomeDB). Paradoxically, autoantibodies to tumor antigens can be found more frequently in patients with missense mutations than in patients with nonsense mutations although nonsense mutations such as stop, splice/stop, splice, or frameshift mutations encode more severely altered proteins (37). One explanation may be that the RNA quality control mechanism NMD allows eukaryotic cells to recognize and degrade mRNAs which contain a nonsense mutation. Consequently, this elimination of mRNAs containing nonsense mutations helps to protect cells against harmful dominant negative effects which could be caused by the encoded truncated mutant protein if translated. In addition, the absence of a translation product of the mutant mRNA may protect the mutant cell against its detection and destruction by the immune system.
In accord with this reasoning, we expected that the identified frame shift mutation in the patient’s La cDNA should not result in an immune response. However, we detected anti-NeoLa Abs in the serum of this patient and even more surprising in about 30% of all analyzed anti-La positive autoimmune patients. Anti-NeoLa Abs were detected with two independent techniques. Using the immunoblotting technique, one cannot identify anti-NeoLa Abs in serum samples which react with the native N-domain of La protein. Consequently, the immunoblotting technique results in an underestimation of the anti-NeoLa frequency. However, the ELISA specific for the NeoLa peptide allowed us to overcome this limitation.
At present, we do not know whether the occurrence of the mutant La was the event for triggering of the anti-NeoLa immune response in the autoimmune patient(s) or the mutant La epitope became a target of an (auto)immune response because it was expressed just at the time when the immune response occurred. At least in the mutant La transgenic mice, which should be tolerant to the mutant La form, the latter mechanism seems to work. In patients who express the mutant La epitope after establishing tolerance both possibilities or a combination of them could also be considered.
Either way, the development of an IgG type anti-NeoLa response requires the expression of mutant La or a mutant La related epitope at the time when the immune response started. One prerequisite for the expression of mutant La protein, however, is an escape of the mutant La mRNA from RNA control. In a previous study, we had shown that E. coli and insect cells can partially correct the mutant La reading frame by ribosomal frame shifting. Ribosomal frame shifting most likely occurs in the oligo(A)-region upstream of the PTC in exon7 (34). Due to this ribosomal frame shifting, expression of the mutant La mRNA does not only result in the expression of the prematurely terminated truncated form of La. In addition to the mutant La protein, a portion of correct human La was made. However, whether this ribosomal frame shifting could also occur in mammalian cells was unclear. If so, the ribosomes translating the mutant La mRNA have to slip into the native La reading frame. The shift has to occur upstream of the PTC and, thus, the ribosomes will bypass the PTC. During translation of the native reading frame downstream of the PTC, these ribosomes can pass all exon splice junctions downstream of the PTC in the mutant La mRNA and terminate translation at the native La mRNA stop codon. Thereby, all EJCs downstream of the PTC will be removed. Consequently, NMD cannot detect the frame shift mutation in the mutant La mRNA, and the mutant La mRNA will, thus, circumvent NMD.
In order to experimentally verify that mutant La mRNA can escape from NMD by ribosomal frame shifting, we established two independent mouse lines transgenic for mutant La. Southern blotting verified the integration of the respective transgene into independent genomic regions in these two lines of mice (data not shown). Quantitative PCR, immunoblotting analysis, and immunohistology show that the mutant La mRNA is not eliminated by NMD. In addition to mutant La, a protein is made which (i) has the same size as native human La protein, (ii) was coprecipitated by patient anti-La antibodies, and (iii) reacted with three mAbs on immunoblots. The used anti-La mAbs recognize unrelated epitopes in either the N- or the C-domain of La. Therefore, the anti-La reactive protein contains La epitopes upstream and downstream of the PTC in the mutant La mRNA. This strongly supports the interpretation that ribosomal frame shifting upstream of the PTC in mutant La mRNA is not restricted to bacterial and insect cells but can also occur in mammalian cells.
In summary, the capability to correct the mutant La reading frame into the native La reading frame by ribosomal frame shifting may be the reason for escape of mutant La mRNAs from NMD.
Both mouse lines develop anti-nuclear Abs including to dsDNA and an immune complex nephritis. The observation of a lupus phenotype in both transgenic lines strongly argues that this phenotype is critically dependent upon the mutant La transgene itself. Thus, expression of mutant La seems to result in an increased risk for developing an autoimmune phenotype.
As La protein is hypothesized to be involved in so many different cellular functions, there are several possibilities as to how the expression of mutant La enhances the risk of an autoimmune response in the transgenic mice. For example, La protein was hypothesized to work as a RNA chaperone and to be involved in the correct folding of RNPs including for example in RNA quality control of 5S ribosomal RNA. An impaired assembly of ribosomes and/or an increased rate of incorrectly folded RNPs could easily lead to neoepitopes which could become the target of an immune response. Alternatively, the mutant La protein could indirectly interfere with the apoptosis pathway allowing perhaps the survival of autoreactive T- and B-cells (31,32). Many recently described transgenic or knock out mice which interfere with the expression of components of the immune system or the apoptosis pathway develop autoimmune phenotypes (38). Although a direct link between La protein, apoptosis resistance, and the immune system has not yet been experimentally shown, there are reports supporting such a function. As mentioned in the introduction section La protein was shown to be involved in the cap-independent translation of the X-linked inhibitor of apoptosis. Moreover, La protein is involved in the translation of the mdm2 protein (21,22). The mdm2 protein is a negative regulator of p53. p53 is a key regulator in DNA repair, the cell cycle and the apoptosis pathway. E.g., p53 is a transcription factor of the cell cycle inhibitor p21. Consequently, a downregulation of mdm2 would result in an upregulation of p21. Interestingly, p21 is known to be upregulated in many experimental autoimmune animal models, and is assumed to stabilize autoreactive memory cells in SLE patients (39). Consequently, an altered expression of La protein could affect the apoptosis resistance and the survival of autoreactive immune cells. Indeed, preliminary data show that La overexpressing cells have an increased apoptosis resistance while knocking down the expression of La increases the apoptosis sensitivity of cells (Bachmann, unpublished). Furthermore, realtime PCR shows a 100-fold upregulation of p21 including in thymus in the mutant La transgenic mice but not in control animals (Bachmann, unpublished).
In summary, our data suggest that mutant La mRNAs can circumvent NMD most likely via ribosomal frame shifting. Expression of mutant La can have two effects (i) it can become a target of an immune response in patients and mutant La transgenic mice, and (ii) it increases the risk of developing systemic autoimmunity, at least in mutant La transgenic mice. The expression of mutant La may interfere with one or all of the functions of the La antigen which finally leads to a predisposition towards an autoimmune response. In contrast to the occurrence of autoimmunity in many other transgenic mice, the autoimmune response in the mutant La transgenic mice may reflect a true risk factor in human SLE, as suggested by the escape mechanism described here and as suggested by the detection of Abs to the NeoLa form in a significant fraction of SLE patients.
Acknowledgments
This work was supported by grants to: MB (GM63497 by the NIH (National Institutes of Health), and HR01-003 by OCAST (Oklahoma Center for the Advancement of Science and Technology OK, U.S.A.), Fritz-Thyssen Stiftung (Germany) and BMBF (Bundesministerium für Forschung und Technik, Germany), Dr. Capra/MB (IP20RRI5577 by the NIH), JBH (AI31584, DE015223, AI053747, AI054117, AR049084, AR48940, AR42460, AI24717, and RR020143 by the NIH), DF (AI51647, AI48097, and AR48940, by the NIH). MB was a recipient of a Greenberg-Scholarship (ORMF, OK, U.S.A.).
Abbreviations
- EJC
exon junction complex
- NMD
nonsense mediated decay of mRNA
- PTC
premature termination codon
- nLa
native human La
- muLa
mutant human La
- moLa
native mouse La
- NeoLa
a neoepitope specific for muLa
- MTC
multi-tissue-cDNA panel
Footnotes
Publisher's Disclaimer: This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This version of the manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence, it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the U.S. National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org.
References
- 1.Fasken MB, Corbett HA. Process or perish: quality control in mRNA biogenesis. Nature Struct Biol. 2005;12:482–489. doi: 10.1038/nsmb945. [DOI] [PubMed] [Google Scholar]
- 2.Weischenfeldt J, Lykke-Andersen J, Porse B. Messenger RNA Surveillance: Neutralizing Natural Nonsense. Current Biol. 2005;15:R559–R562. doi: 10.1016/j.cub.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Schell T, Kocher T, Wilm M, Seraphin B, Kulozik AE, Hentze MW. Complexes between the nonsense-mediated mRNA decay pathway factor upf1 (up-frameshift protein 1) and essential nonsense-mediated mRNA decay factors in HeLa cells. Biochem J. 2003;373:775–783. doi: 10.1042/BJ20021920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Culbertson MR. RNA surveillance: unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 1999;15:74–80. doi: 10.1016/s0168-9525(98)01658-8. [DOI] [PubMed] [Google Scholar]
- 5.Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 6.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 7.Tröster H, Metzger T, Semsei I, Schwemmle M, Winterpacht A, Zabel M, Bachmann M. One Gene, two transcripts: Isolation of an alternative transcript encoding for the autoantigen La/SS-B from a cDNA library of a patient with primary Sjögren’s syndrome. J Exp Med. 1994;180:2059–2067. doi: 10.1084/jem.180.6.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grölz D, Laubinger J, Wilmer F, Tröster H, Bachmann M. Transfection analysis of expression of mRNA isoforms encoding the nuclear autoantigen La/SS-B. J Biol Chem. 1997;18:12076–12082. doi: 10.1074/jbc.272.18.12076. [DOI] [PubMed] [Google Scholar]
- 9.Grölz D, Bartsch H, Tröster H, Bachmann M. The nuclear autoantigen La/SS-B: Mapping and sequencing of the gene and the three retropseudogenes. Gene. 1997;191:23–29. doi: 10.1016/s0378-1119(97)00023-1. [DOI] [PubMed] [Google Scholar]
- 10.Bachmann M, Tröster H, Bartsch H, Grölz D. A frame shift mutation in a hot spot of the nuclear autoantigen La/SSB. J Autoimmun. 1996;9:747–756. doi: 10.1006/jaut.1996.0097. [DOI] [PubMed] [Google Scholar]
- 11.Simons FH, Broers FJ, Van Venrooij WJ, Pruijn GJ. Characterization of cis-Acting Signals for nuclear import and retention of the La (SS-B) autoantigen. Exp Cell Res. 1996;224:224–236. doi: 10.1006/excr.1996.0132. [DOI] [PubMed] [Google Scholar]
- 12.Maraia RJ, Kenan DJ, Keene JD. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol Cell Biol. 1994;14:2147–2158. doi: 10.1128/mcb.14.3.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maraia RJ. Transcription termination factor La is also an initiation factor for RNA polymerase III. Proc Natl Acad Sci USA. 1996;93:3383–3387. doi: 10.1073/pnas.93.8.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan H, Sakulich AL, Goodier JL, Zhang X, Qin J, Maraia RJ. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell. 1997;88:707–715. doi: 10.1016/s0092-8674(00)81913-3. [DOI] [PubMed] [Google Scholar]
- 15.Yoo CJ, Wolin SL. The Yeast La Protein Is Required for the 3′Endonucleolytic Cleavage That Matures tRNA Precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]
- 16.Gottlieb E, Steitz JA. The RNA binding protein La influences both the accuracy and the efficiency of RNA polymerase III transcription in vitro. EMBO J. 1989;8:841–850. doi: 10.1002/j.1460-2075.1989.tb03445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottlieb E, Steitz JA. Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J. 1989;8:851–861. doi: 10.1002/j.1460-2075.1989.tb03446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLaren RS, Caruccio N, Ross J. Human La Protein: a Stabilizer of Histone mRNA. Mol Cell Biol. 1997;17:3028–3036. doi: 10.1128/mcb.17.6.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meerovitch K, Pelletier J, Sonenberg N. A cellular protein that binds to the 5′-non-coding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 1989;3:1026–1034. doi: 10.1101/gad.3.7.1026. [DOI] [PubMed] [Google Scholar]
- 20.Meerovitch K, Svitkin YV, Lee HS, Lejbkowicz F, Kenan DJ, Chan EK, Agol VI, Keene JD, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svitkin YV, Meerovitch K, Lee HS, Dholakia JN, Kenan DJ, Agol VI, Sonenberg N. Internal translation initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J Virol. 1994;68:1544–1550. doi: 10.1128/jvi.68.3.1544-1550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svitkin YV, Pause A, Sonenberg N. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J Virol. 1994;68:7001–7007. doi: 10.1128/jvi.68.11.7001-7007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutierrez-Escolano L, del Angel RM. Nuclear proteins bind to poliovirus 5′ untranslated region. Arch Med Res. 1996;27:413–419. [PubMed] [Google Scholar]
- 24.Ali N, Siddiqui A. The La Antigen binds 5′noncoding region of the Hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pogue GP, Hofmann J, Duncan R, Best JM, Etherington J, Sontheimer RD, Nakhasi HL. Autoantigens Interact with cis-Acting Elements of Rubella Virus RNA. J Virol. 1996;70:6269–6277. doi: 10.1128/jvi.70.9.6269-6277.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De BP, Gupta S, Zhao H, Drazba JA, Banerjee AK. Specific Interaction in Vitro and in Vivo of Glyceraldehyde-3-phosphate Dehydrogenase and La Protein with Cis-acting RNAs of Human Parainfluenza Virus Type 3. J Biol Chem. 1996;271:24728–24735. doi: 10.1074/jbc.271.40.24728. [DOI] [PubMed] [Google Scholar]
- 27.Park YW, Katze MG. Translational Control by Influenza Virus. J Biol Chem. 1995;270:28433–28439. doi: 10.1074/jbc.270.47.28433. [DOI] [PubMed] [Google Scholar]
- 28.Jefferies HB, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pellizzoni L, Cardinali B, Lin-Marq N, Mercanti D, Pierandrei-Amaldi P. Xenopus laevis Homologue of the La autoantigen binds the pyrimidine tract of the 5′ UTR of ribosomal protein mRNAs in vitro: Implication of a protein factor in complex formation. J Mol Biol. 1996;259:904–915. doi: 10.1006/jmbi.1996.0368. [DOI] [PubMed] [Google Scholar]
- 30.Perrotti D, Calabretta B. Translational regulation by the p210 BCR/ABL oncoprotein. Oncogene. 2004;23:3222–3229. doi: 10.1038/sj.onc.1207543. [DOI] [PubMed] [Google Scholar]
- 31.Holcik M. Translational upregulation of the X-linked inhibitor of apoptosis. Ann N Y Acad Sci. 2003;1010:249–258. doi: 10.1196/annals.1299.043. [DOI] [PubMed] [Google Scholar]
- 32.Trotta R, Vignudelli T, Candini O, Intine RV, Pecorari L, Guerzoni C, Santilli G, Byrom MW, Goldoni S, Ford LP, Caligiuri MA, Maraia RJ, Perrotti D, Calabretta B. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell. 2003;3:145–160. doi: 10.1016/s1535-6108(03)00020-5. [DOI] [PubMed] [Google Scholar]
- 33.Buyon JP, Hiebert R, Copel J, Craft J, Friedman D, Katholi M, Lee LA, Provost TT, Reichlin M, Rider L, Rupel A, Saleeb S, Weston WL, Skovron ML. Autoimmune-associated congenital heart block: Mortality, morbidity, and recurrence rates obtained from a national neonatal lupus registry. J Am Coll Cardiol. 1998;31:1658–1666. doi: 10.1016/s0735-1097(98)00161-2. [DOI] [PubMed] [Google Scholar]
- 34.Bachmann M, Grölz D, Bartsch H, Klein RR, Tröster H. Analysis of expression of an alternative La (SS-B) cDNA and localization of the encoded N- and C-terminal peptides. Biochim Biophys Acta. 1997;27:53–63. doi: 10.1016/s0167-4889(96)00158-9. [DOI] [PubMed] [Google Scholar]
- 35.Keech CL, Farris AA, Beroukas D, Gordon TP, McCluskey J. Cognate T cell help is sufficient to trigger anti-nuclear autoantibodies in naive mice. J Immunol. 2001;166:5826–5835. doi: 10.4049/jimmunol.166.9.5826. [DOI] [PubMed] [Google Scholar]
- 36.Sahin U, Türeci Ö, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci USA. 1995;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winter SF, Minna JD, Johnson BE, Takahashi T, Gazdar AF, Carbone DP. Development of Abs against p53 in lung cancer patients appears to be dependent on the type of p53 mutations. Cancer Res. 1992;52:4168–4174. [PubMed] [Google Scholar]
- 38.Wakeland EK, Liu K, Graham RR, Behrens TW. Delineating the genetic basis of systemic lupus erythematosus. Immunity. 2001;15:397–408. doi: 10.1016/s1074-7613(01)00201-1. [DOI] [PubMed] [Google Scholar]
- 39.Lawson BR, Baccala R, Croft M, Kono DH, Theofilopoulos AN. Deficiency of cyclin kinase inhibitor p21(WAF-1/CIP-1) promotes apoptosis of activated/memory T-cells and inhibits spontaneous systemic autoimmunity. J Exp Med. 2004;199:547–557. doi: 10.1084/jem.20031685. [DOI] [PMC free article] [PubMed] [Google Scholar]