Figure 2.
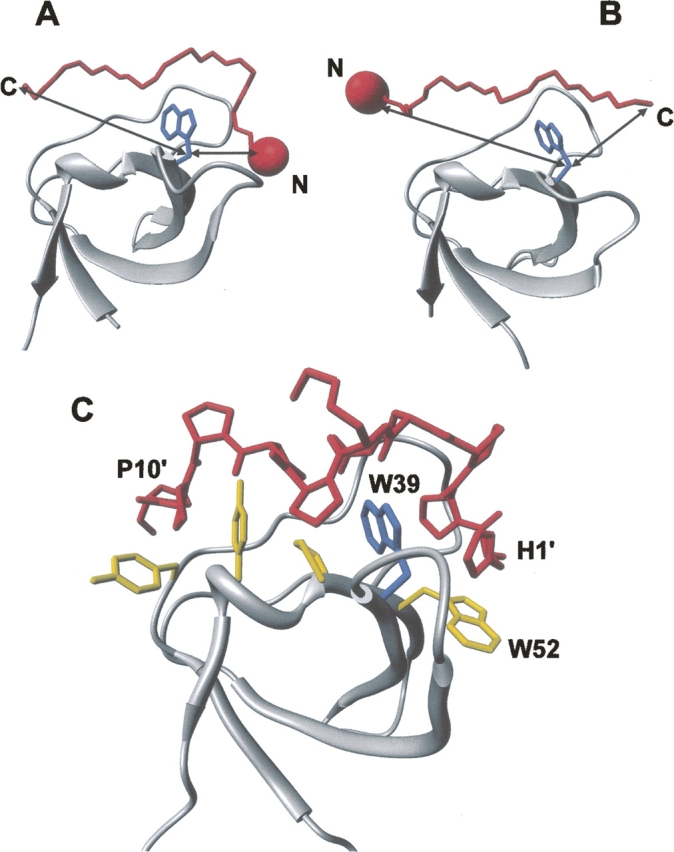
Structural analysis of SH3 complexes to illustrate the strategy proposed. Structures of two representative SH3 domains (ribbon drawing, gray) bound to (A) class I (1abo) (Musacchio et al. 1994) and (B) class II (1cka) (Wu et al. 1995) peptides (Cα trace, red). (Blue) The side chain of the conserved Trp residue in the SH3 domain; (arrows) the distances (Cβ–Cβ atoms) between the peptide termini and Trp. The incorporation of a suitable spectroscopic probe (red sphere), like NT or IF, is expected to perturb the intrinsic fluorescence of the conserved Trp in the SH3 domain, in a way that is strongly dependent on the orientation of the binding peptide on the SH3 surface. For instance, if the probe is incorporated at the N terminus of the ligand peptide, the fluorescence of SH3 is expected to be quenched only by a class I peptide, whereas it should be essentially unaffected if the ligand is a class II peptide. (C) Structure of the P2/Myo3-SH3 complex (Musi et al. 2006), showing relevant interactions of the ligand peptide (stick, red) with the SH3 domain (ribbon drawing, gray). The side chains of key amino acids of Myo3-SH3 are also explicitly shown (stick, yellow), while the conserved Trp39 is shown in blue.
