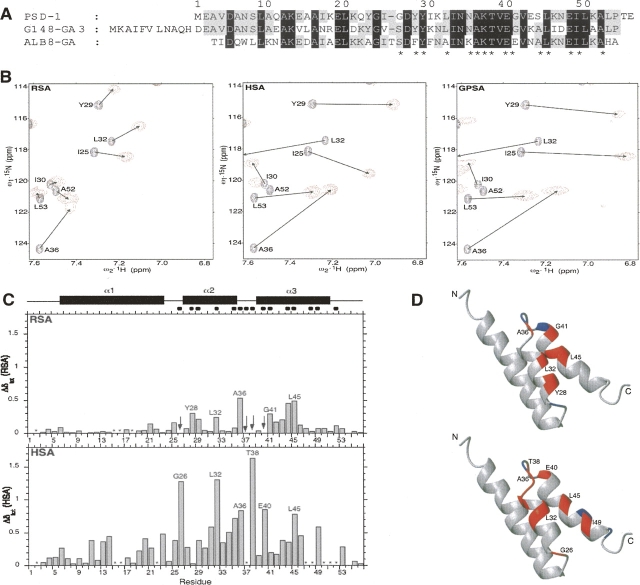
Figure 1.
(A) Alignment of PSD-1, streptococcal G148–GA3, and Finegoldia magna ALB8–GA albumin-binding domains. Numbering is for PSD-1 residues. Identical residues are in gray or black, corresponding to conservation across two or three sequences, respectively. Amino acids implicated at the albumin-binding interface according to the ALB8–GA/HSA X-ray structure are indicated with asterisks. (B) Sections from 15N HSQC spectra of albumins complexed with [15N]-PSD-1. Cross-peaks due to free PSD-1 are black and those due to albumin-complexed PSD-1 are red. (C) Histogram plots of chemical shift perturbation for PSD-1 with RSA (top) and HSA (bottom). The secondary structure regions (filled rectangles) and albumin binding epitope (filled circles) are indicated at the top of the figure. The epitope is based on corresponding GA module residues contacting albumin in the X-ray structure of the ALB8–GA/HSA complex (Lejon et al. 2004). Asterisks indicate positions for which Δδ could not be determined unambiguously due to overlapping or weak signals in the complex. Arrows indicate exchange broadened signals. (D) Ribbon representations of the uncomplexed PSD-1 NMR structure (He et al. 2006) are used to highlight RSA (top) and HSA (bottom) complexed PSD-1 residues with significant Δδ in red. Epitope residues for which Δδ could not be determined unambiguously are rendered in blue.