Abstract
Sm-like proteins are ubiquitous ring-shaped oligomers that exhibit a variety of nucleic acid-binding activities. They have been linked functionally to various cellular events involving RNA, and it is generally believed that their activity is exerted via the passive binding of nucleic acids. Our earlier studies of the Sm-like Escherichia coli protein Hfq provided the first evidence indicating that Hfq is an ATP-binding protein. Using a combination of biochemical and genetic techniques, we have now determined a plausible ATP-binding site in Hfq and tested Hfq's ATP-binding affinity and stoichiometry. The results of RNA footprinting and binding analyses suggest that ATP binding by the Hfq–RNA complex results in its significant destabilization. RNA footprinting indicates deprotection of Hfq-bound RNA tracts in the presence of ATP, suggestive of their release by the protein. The results reported herein broaden the scope of potential in vivo roles for Hfq and other Sm-like proteins.
Keywords: Hfq, Sm, Sm-like, translation, conformational change, electrophoresis
Sm and Sm-like proteins are ubiquitous, phylogenetically well-conserved ring-shaped oligomers that display nucleic acid-binding activity. Functionally, these proteins have been linked to various cellular events that involve RNA, such as translation, RNA splicing, RNA degradation, and other forms of riboregulation (for review, see Pannone and Wolin 2000), and it is generally believed that the activity of Sm- and Sm-like proteins is exerted via non-energy-dependent and/or cofactor-regulated protein–nucleic acid interactions. Structurally, the characteristic feature of Sm- and Sm-like proteins—and a basis for their classification as a single protein family—is an OB-like fold, consisting of five bent antiparallel β-sheets capped by an N-terminal α-helix.
Bacterial Hfq proteins—the Escherichia coli representative of which was characterized early on as a Host cell factor essential for the replication of the q-β bacteriophage (Franze de Fernandez et al. 1972)—are a family of hexameric nucleic acid-binding proteins, whose homology with Sm-like proteins was initially suggested by sequence similarity among their N-terminal segments. Identification of Hfq as the bacterial branch of the Sm family has been further corroborated by phylogenetic, physico-chemical, homology modeling, and crystallographic studies (Arluison et al. 2002, 2006; Moller et al. 2002; Schumacher et al. 2002; Sun et al. 2002; Zhang et al. 2002; Nikulin et al. 2005).
Biochemical studies of Hfq demonstrated its RNA-binding activity in vitro, which was confirmed with a variety of RNAs, including small stable RNAs (Vytvytska et al. 1998; Sledjeski et al. 2001; Moller et al. 2002; Schumacher et al. 2002). Some studies have demonstrated the preferential binding of Hfq to adenine- and uridine-rich tracts in RNA (Folichon et al. 2003; Moll et al. 2003). In the X-ray structure of the Hfq–RNA complex, the RNA oligonucleotide is bound around the inner rim of the Hfq doughnut, interacting with multiple individual subunits (Schumacher et al. 2002). Furthermore, recent studies have suggested that Hfq may possess additional RNA-binding sites (Brescia et al. 2003; Mikulecky et al. 2004; Sun and Wartell 2006). A recent study showed that, like other Sm proteins, individual Hfq hexamers can self-associate into well-ordered helical fibers (Arluison et al. 2006).
Information regarding the subcellular localization of Hfq, which is critical for a thorough understanding of protein function, is more scarce. Early on, it was suggested that Hfq interacts with ribosomes via the 30S ribosomal subunit (DuBow et al. 1977). Consistent with this, Hfq was shown to copurify in stoichiometric amounts (1:1) with the ribosomal protein S1, which has been implicated in a specific interaction with the 5′-termini of mRNAs (e.g., see Sengupta et al. 2001), and to interact with RNA polymerase in an S1-dependent manner (Sukhodolets and Garges 2003). Studies carried out to identify the subcellular localization of Hfq found that most of the protein is associated with the translational machinery, and a minor fraction appears to be associated with the nucleoid (Kajitani et al. 1994; Azam et al. 2000), possibly via nucleoid-associated polysomes.
In vivo studies have shown that inactivation of the hfq gene in E. coli causes a variety of phenotypes and alters the expression of many proteins (Tsui et al. 1994; Muffler et al. 1997). Hfq was demonstrated to be important for the stability of several cellular RNAs, including riboregulators involved in bacterial quorum sensing (Lenz et al. 2004; Mohanty et al. 2004), and it was shown that several mRNAs are controlled by the concerted action of regulatory RNAs and Hfq (for review, see Wassarman 2002; Masse et al. 2003). Hfq was reported to target some mRNAs for degradation by binding to their poly(A) tails and stimulating their polyadenylation (for review and a complete set of references, see Valentin-Hansen et al. 2004). On this basis, many studies have described Hfq as a “pleiotropic regulator” (Tsui et al. 1996, 1997; Schumacher et al. 2002). It was suggested that the physiological activity of Hfq may be explained by the protein's passive modulation of RNA–RNA interactions (Moller et al. 2002; Zhang et al. 2002). Consequently, recent studies have described Hfq as an omnipresent protein regulator rather than an element of the translational machinery with a narrowly defined function (e.g., see Valentin-Hansen et al. 2004 and references therein). The consensus from most of these studies is the proposed role of Hfq as an RNA chaperone.
Primarily through biochemical means, our previous study identified Hfq as the source of the E. coli RNA polymerase-associated latent ATPase activity (Sukhodolets and Garges 2003)—a surprising finding given the protein's small size and ostensible lack of definitive sequence homology with known ATP-utilizing enzymes. In the present study, based on additional biochemical and genetic results, including identification of the Hfq ATPase near-knockout Tyr25Ala mutation, we have determined a plausible location for the Hfq ATP-binding site. Furthermore, our biochemical and electron microscopic data show a significant effect of ATP on the structure and stability of the Hfq–RNA complex. Taken together, our data indicate that the specific functions of Sm-like Hfq proteins thus far established in purified systems should be reexamined in light of the possible role of ATP in Hfq-mediated RNA remodeling.
Results
Mutational analysis of the Hfq ATPase
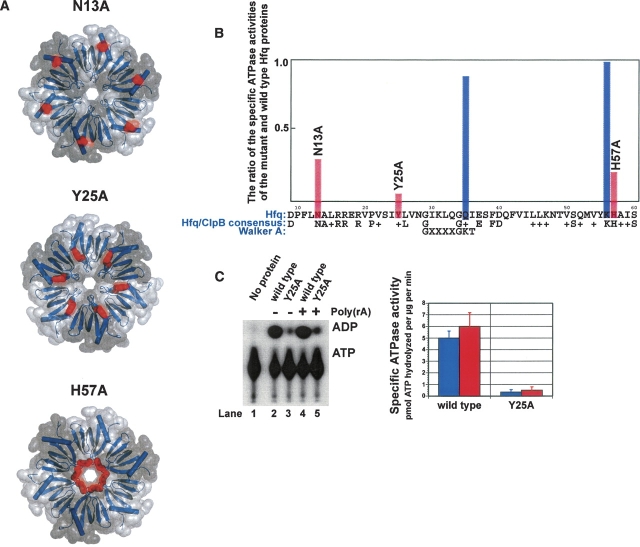
We first expanded on our previous strategy of constructing mutant Hfq proteins and testing their specific ATPase activity in vitro in order to determine how individual mutations affect the latent ATP hydrolysis that is observed in our Hfq preparations (Sukhodolets and Garges 2003). In our previous set of mutational analyses, we selected amino acid sites based primarily on the degree of conservation of these individual sites among E. coli Hfq and its homologs. However, mutations in the Hfq equivalent of the Walker A box and the mutation of Lys56 (a highly conserved residue in Hfq proteins) produced only marginal alterations in the efficiency of ATP hydrolysis (Sukhodolets and Garges 2003). In a new set of experiments, we surveyed the Hfq structure and mutagenized three individual amino acids located at topologically distinct sites, namely, His57 (which, with the other five histidines in the Hfq hexamer, lines the entrance to the pore), Tyr25 (which flanks six distinct intersubunit pockets located on the “L4” face [adopting the nomenclature used in Arluison et al. 2006, which was initially suggested in Mura et al. 2003] of the Hfq disc), and Asn13 (a conserved amino acid near the outer rim of the Hfq ring) (▶). The results of this set of experiments are summarized in ▶. Alanine substitution of His57 produced a modest alteration in ATP hydrolysis, and the Asn13Ala mutation had only a marginal effect on the specific ATPase activity of purified Hfq (▶). In contrast, the Tyr25Ala mutation resulted in a near-knockout of ATP hydrolysis in the purified protein (▶).
Figure 1.
The effect of site-specific mutations on the efficiency of ATP hydrolysis by purified Hfq proteins. (A) The location of the N13A, Y25A, and H57A mutation sites (shown in red) in the Hfq molecule. Note that the N13A and H57A panels are viewed looking onto the “bottom” of the disc-shaped hexamer (i.e., N-terminal helical face), whereas the Y25A illustration is viewed looking onto the Loop-4 (“L4”) face of the hexamer. (B) For each of the indicated mutations, parallel purifications of the wild-type and mutant Hfq proteins were carried out, and the Hfq-specific ATPase activities were determined as described previously (Sukhodolets and Garges 2003; see page 8023 therein). The ratios of the two specific activities are plotted versus the Hfq amino acid sequence. Data represent the average of two or more independent experiments. (Blue columns) Previously constructed mutations (Sukhodolets and Garges 2003); (red columns) mutations described in this study. (C) A representative PEI-cellulose plate on which the activities of the Hfq near-knockout Y25A mutant protein and the wild-type Hfq protein are tested side by side. The quantitated results of this experiment are shown at right; reactions were performed in the (blue columns) absence or in the (red columns) presence of excess Poly(rA). Data represent the average of two independent experiments.
RNA and nucleotide binding by wild-type Hfq and the Hfq Tyr25Ala mutant
To characterize the Hfq ATPase near-knockout mutant Tyr25Ala, we tested the ATP- and RNA-binding properties of the mutant protein in comparison to those of wild-type Hfq. First, we analyzed the RNA-binding affinities of the two proteins by monitoring changes in the fluorescence anisotropy of 5′-fluoresceinated RNA oligonucleotides in the presence of Hfq. For wild-type Hfq, the equilibrium dissociation constant (K d) for rA7 was 24 ± 3 nM (▶), whereas the Tyr25Ala Hfq mutant showed a reduced affinity for rA7 (K d = 118 ± 7 nM) (▶). Reduced RNA binding resulting from the Hfq Y25A mutation (albeit in protein obtained from a dissimilar purification procedure; see Materials and Methods) has been reported (Mikulecky et al. 2004; Sun and Wartell 2006). The interaction of Hfq with rA7 was selective, since no interaction with rC7 and dA7 was detected in this binding assay, even at Hfq concentrations exceeding 1 μM.
Figure 2.
The effect of ATP on Hfq–RNA interaction. Experiments using 5′(fluorescein)-modified RNA probes were carried out as described in Materials and Methods. (A) ATP diminishes the RNA-binding activity of Hfq. Titrations of rA7 in the (rectangles) absence or in the (triangles) presence of 1 mM AMP-PNP, (rhombuses) 1 μM ATP, or (circles) 1 mM ATP with wild-type Hfq produced K d values of 27, 28, 30, and 100 nM, respectively. The decrease in fluorescence anisotropy in the presence of excess ATP is consistent with our central claim that ATP modulates RNA–Hfq interactions; we believe that the anisotropy change may reflect complex interactions between RNA-fluorophore and the Hfq hexamer in an altered conformation. (B) Titration of rA7 with Y25A Hfq resulted in the determination of a K d value of 118 nM.
We used a similar approach for comparison of the nucleotide-binding affinities of wild-type Hfq and Tyr25Ala Hfq. In these experiments, the changes in fluorescence anisotropy of 2′/3′-O-(N-methyl-antraniloyl) nucleotides (MANT nucleotides) in the presence of Hfq were monitored. Before the affinity measurements, stoichiometric titration of Hfq with the nucleotide was performed in order to determine the number of nucleotide-binding sites (▶). The equivalence point indicates that approximately one nucleotide is bound per Hfq monomer. For wild-type Hfq, K ds were 0.75 ± 0.05 μM, 0.74 ± 0.05 μM, and 0.76 ± 0.05 μM for ADP, ATP, and AMP-PNP, respectively (▶). Similar values were obtained in the presence of 2 mM Mn2+ or Mg2+, suggesting that nucleotide binding by Hfq is unaffected by the nature of the cation in the buffer, unlike the reported ATP hydrolytic activity (Sukhodolets and Garges 2003). This result points to a catalytic role of the cation rather than direct involvement in ATP binding. In the case of the Tyr25Ala Hfq mutant, the K d for ADP was 1.7 ± 0.1 μM; ATP and AMP-PNP showed similar affinities (▶). We also determined the Hfq K d for rA7 in the presence of 1 μM of either ATP, ATP-γ-S, or AMP-PNP. These constants were 30 ± 3, 27 ± 2, and 28 ± 2 nM, respectively (data not shown). Thus, the protein shows comparable RNA-binding affinities in the presence or absence of nucleotides. In contrast, at 1 mM ATP (approximately four K m values, based on the earlier reported Hfq K m for ATP of 0.25 mM) (Sukhodolets and Garges 2003), the K d for rA7 RNA increased to 95 ± 5 nM (▶). Thus, ATP binding at a concentration higher than K m decreases the affinity of Hfq for the RNA oligonucleotide approximately by a factor of 4.
Figure 3.

Determination of the number of ATP-binding sites and the affinities of Hfq for nucleotides. Experiments using MANT-modified nucleotides were carried out as described in Materials and Methods. (A) Titration of MANT-ATP with the Hfq protein. The equivalence point, which was reached at ∼10 μM Hfq monomer (at 10 μM MANT-ATP), gives a binding stoichiometry of 6 ATP molecules per Hfq hexamer. (B) Titrations of (rectangles) MANT-ATP, (triangles) MANT-ADP, and (circles) MANT-AMP-PNP with Hfq produced similar dissociation constants of ∼0.75 μM. (C) Parallel titrations of MANT-ATP with (rectangles) wild-type and (triangles) Y25A Hfq gave dissociation constants of ∼0.75 and 1.75 μM, respectively.
The effect of ATP on Hfq–RNA complexes studied by RNA footprinting and EMSA
Because our binding studies with modified nucleotide and RNA probes revealed a destabilizing effect of ATP on Hfq–RNA complexes, we set out to confirm this effect by alternative techniques, using unmodified RNA probes. First, we carried out footprinting analyses of the Hfq–RNA complexes in the presence or absence of ATP. These experiments used short synthetic RNAs incorporating two 5′-terminal hairpins followed by an 18-nt 3′-terminal rA tail (RNA A; Dharmacon). (RNA A, shown schematically in ▶, mimics the basic structures found in the 5′- and 3′-termini of mRNAs.) With purified wild-type Hfq, the region in RNA A most efficiently protected from ribonuclease digestion was the interface of the rA tail and the adjacent hairpin (▶). This result is consistent with our binding studies using fluorescein-labeled oligonucleotides, as well as other reported data that indicate preferential Hfq binding to adenine-rich tracts in RNA (Folichon et al. 2003; Moll et al. 2003), and the previously reported footprinting analyses of Hfq–RNA complexes (Zhang et al. 2002). Addition of ATP resulted in significant de-protection of most otherwise protected regions (▶, arrows), with subtle changes in the de-protection efficiency at individual sites. The de-protection was most significant at the junction of the rA tail and the adjacent hairpin. In contrast, Tyr25Ala Hfq showed no pronounced RNA A protection, even at concentrations approaching 1 μM (data not shown).
Figure 4.
ATP destabilizes Hfq–RNA complexes. (A) Schematic diagram of the secondary structure of the RNA probe used in these experiments. Key Ribonuclease A cleavage sites are indicated by arrows. (B) Effect of ATP on Hfq binding to a model RNA incorporating a stem–loop structure and 3′-terminal poly(rA) tail studied by Ribonuclease A footprinting. RNA footprinting experiments were carried out as described in Materials and Methods. Reactions in the absence or in the presence of 1 mM ATP-γ-S are shown. The de-protected sites are indicated by arrowheads. (C) Destabilization of the urea-insensitive Hfq–RNA complexes in the presence of ATP. EMSA experiments were carried out as described in Materials and Methods.
Consistent with the results of the footprinting experiments, EMSA-based studies have indicated that the addition of ATP destabilizes high-affinity (urea-insensitive) Hfq–RNA complexes (▶).
Electron microscopic analyses of Hfq in the presence or absence of RNA and nucleotides
We next used electron microscopy to probe the general structural effects of RNA and/or nucleotides on the Hfq hexamer. Rotational power spectra analysis performed on averaged images (▶) verified the expected sixfold symmetry of hexameric Hfq protein alone (▶) and in the presence of rA7 oligonucleotide (▶), ATP (▶), or non-hydrolyzable ATP analogs plus rA7 (▶). This result suggests that ATP binding, per se, does not induce a major structural change in Hfq. In contrast, in the presence of both rA7 and ATP (▶), the sixfold symmetry of the Hfq ring is broken, thereby implying a major structural change. Interestingly, we detected no such change with Tyr25Ala Hfq, suggesting that the mutant protein is unable to undergo this conformational transition seen with wild-type Hfq (▶).
Figure 5.
Disruption of the Hfq sixfold symmetry in the RNA–Hfq–ATP complex. The electron microscopic experiments were carried out as described in Materials and Methods. The lack of the effect in the ternary complexes using non-hydrolyzable ATP analogs suggests that the conformational change accompanying this transition is coupled to ATP hydrolysis.
Functional consequences of the Hfq Tyr25Ala mutation
Next, in order to assess the impact of the Y25A mutation on Hfq function, we tested the effect of this mutation on the efficiency of luciferase translation in Hfq-free S30 extracts. The choice of a coupled transcription–translation system was dictated by our inability to transfer the described hfq deletion mutation from the E. coli MC4100 strain (Tsui et al. 1994; Zhang et al. 2001) into the wild-type (MG1655) background by phage-mediated transduction. Coupled in vitro transcription–translation reactions were carried out under conditions in which the translation step was made rate-limiting by the addition of excess RNA polymerase, as previously described (Sukhodolets and Garges 2003). At 0.2 μM Y25A Hfq, translation of luciferase was suppressed at early time points to a level comparable to that of “minus Hfq” extracts (▶). In contrast, 0.2 μM purified wild-type Hfq showed a moderate stimulatory effect on luciferase translation, consistent with our previously reported results (Sukhodolets and Garges 2003). Interestingly, the S30 extracts exhibited the ability to overcome this transient inhibitory effect of the Hfq Y25A mutant protein after prolonged (>25 min) incubation (▶). Excess Hfq (>2 μM), either the wild-type or Y25A protein, inhibited the coupled transcription–translation reaction (data not shown).
Figure 6.
Functional effects of the Hfq Y25A mutation. Coupled transcription–translation in hfq-minus S30 extracts in the presence of (black columns) 0.2 μM purified wild-type Hfq, (gray columns) 0.2 μM Hfq Y25A, or (open columns) in the absence of Hfq. The coupled in vitro transcription–translation reactions used a pBESTluc™ supercoiled DNA template and were carried out as described (Sukhodolets and Garges 2003).
Taken together, these results support the argument that introducing Y25A Hfq into the in vitro coupled transcription–translation system is moderately disruptive to the E. coli translational apparatus. The effect may be due to reduced ATPase activity and/or RNA binding in the mutant protein; however, the effect of the mutation on Hfq–S1 complexes or complexes of Hfq with other potential interactors cannot be ruled out.
Structural model for the Hfq ATP-binding site
Homology of the C-teminal part of Hfq (which carries the Hfq equivalent of the Walker A-box) to the known ATP-binding protein ClpB and near-knockout of the Hfq ATPase activity following alanine substitution of Tyr25 allowed us to infer possible ATP-binding sites in Hfq (▶). It seems likely that Tyr25 plays an important role in stabilizing the adenine portion of ATP, possibly via aromatic stacking (▶). The cleft flanked by Tyr25 is of appropriate size for accommodating an adenine. Manual docking of ATP into the predicted binding cleft does not allow determination of the exact position of the ATP triphosphate portion (▶), but does suggest plausible models (▶). Additional genetic, biochemical, and molecular modeling studies are currently being performed in order to elucidate the exact nature of ATP binding.
Figure 7.

Structural model for the Hfq ATP-binding site. A. The Hfq hexamer is viewed onto the “L4” face, with protein chains represented as ribbon cartoons. Semitransparent solvent-accessible surfaces are drawn in alternating gray intensities, and an individual Hfq monomer is delimited by dashed lines (β-strands and termini are indicated for this subunit). ATP (green) and Y25 (tan) are rendered as space-filling CPK spheres, and the ATP-binding cleft is shown in more detail in B. Surface colors for residues within 4.5 Å of ATP are graded from (yellow) least conserved to (orange) moderately conserved and (red) most strictly conserved amino acids. (C) In addition to favorable π···π stacking interactions, numerous other ionic and hydrogen-bond contacts are indicated, with the width of the dashed lines (magenta) that represent the interaction scaled by the degree of phylogenetic conservation of that amino acid. The phosphoester tail for alternative, less energetically favorable configurations of bound ATP are drawn as thin lines. Note that the binding pocket lies along the boundary between individual Hfq subunits, and that strongly conserved residues contribute to form a composite binding site capable of providing a network of Hfq···ATP interactions to staple the nucleotide into this surface pocket.
Discussion
The results of this study, taken together with our previous study (Sukhodolets and Garges 2003), establish Hfq as an ATP-binding protein. To the combined body of previously reported evidence implicating Hfq in ATP binding (Sukhodolets and Garges 2003), this study adds the evidence of stoichiometric ATP binding by Hfq (which rules out ATP binding by potential Hfq-associated impurity proteins) and near knockout of the latent Hfq-associated ATPase activity following alanine substitution of Tyr25. In addition, our modeling work shows the overall compatibility of the predicted ATP-binding cleft—flanked by Tyr25—with the configuration of an ATP molecule. Importantly, key amino acids forming the putative ATP-binding clefts (Tyr25 and Gln52) are highly conserved in the bacterial Hfq proteins; and Lys31 (the residue located within the Hfq equivalent of a Walker A-box) is moderately conserved (▶). If the general location of the predicted ATP-binding cleft in Hfq is correct and the triphosphate portion of the ATP molecule does, indeed, point to Hfq's primary catalytic site, this site would be located near the outer rim, at the “L4” face of the doughnut-shaped Hfq molecule.
There was a significant, eight- to 20-fold reduction of the specific ATPase activity in purified Hfq resulting from the Tyr25Ala mutation. In general, the observed rate of ATP hydrolysis by wild-type Hfq in the presence of S1 (as likely pertains to the in vivo context), which exerts a two- to threefold stimulatory effect on the Hfq ATPase (Sukhodolets and Garges 2003), 10–18 pmol of ATP hydrolyzed per microgram of protein per minute, compares favorably with the rate of ATP hydrolysis observed for DnaK, another latent, abundant E. coli ATPase (Zylicz et al. 1983) (15–20 nmol of ATP hydrolyzed per milligram of protein per minute); the two rates are of the same order of magnitude if calculated per Hfq monomer and roughly comparable if calculated per Hfq hexamer.
Titrations of MANT-modified nucleotides with Hfq have confirmed ATP binding by Hfq. The K d values of the Hfq–nucleotide complexes were in the 0.5–1 μM range. All values are in the physiological range for ATP. The binding stoichiometry was ∼6 mol of nucleotide per Hfq hexamer, as expected, and consistent with the number of predicted ATP-binding sites in the Hfq molecule. As in most protein–ligand binding studies that rely on ligands with covalently bound fluorophores, contribution of the fluorophore to the interaction cannot be ruled out entirely. However, owing to (1) our controls indicating a competition on the part of unlabeled ATP and RNA with labeled probes for binding to Hfq (see Materials and Methods) and, more importantly, (2) the fact that EMSA and footprinting experiments—which used unmodified nucleotide and RNA probes—also imply ATP–Hfq interaction (since it is highly unlikely that the destabilizing effects of ATP on Hfq–RNA complexes observed in the two latter types of assays are due to ATP–RNA interaction), we believe that it is unlikely that either fluorophore is a major contributor to the interaction.
Our electron microscopy work provides evidence that the ternary RNA–Hfq–ATP complex is structurally distinct from the RNA–Hfq, Hfq–nucleotide, or RNA–Hfq–non-hydrolyzable ATP analog complexes. The data indicate that the binding of both RNA and ATP is accompanied by a detectable distortion in the Hfq sixfold symmetry. This result is consistent with a change in fluorescence anisotropy in Hfq–RNA complexes in the presence of ATP, as shape influences rotational motions. Such an alteration would not be possible without a significant conformational change in Hfq. Even though the exact nature of this conformational change remains to be determined, our data may point to one of its functional consequences. Independently, the binding studies with 5′-fluoresceinated RNAs, RNA footprinting, and EMSA experiments (the latter techniques used unmodified RNA probes) demonstrated destabilization of the Hfq–RNA complexes in the presence of ATP. The fluorescence-based binding studies demonstrated an approximately fourfold reduction in Hfq RNA-binding affinity for rA7 in the presence of excess ATP. (While these experiments, taken separately, may allow multiple interpretations of the reduced fluorescence anisotropy in the presence of ATP, taken together with the results of EMSA and ribonuclease A footprinting experiments, the effect in question is more likely due to destabilization of the Hfq–RNA complex, which may manifest itself in either dissociation or partial peeling of RNA from the protein in the presence of excess ATP.) The RNA footprinting analyses with longer RNA probes—which allowed for greater precision in monitoring the protein–RNA interactions at specific sites in the RNA—indicated significant de-protection of particular protein-bound RNA tracts in the presence of ATP, indicative of their release by the protein. Independently, PAGE-based binding assays have produced similar results, indicating that ATP destabilizes the Hfq–RNA complex significantly. These findings challenge the commonly accepted role of Hfq as a protein that exerts its activity in vivo via high-affinity, passive RNA binding. Nearly all reported in vitro studies with Hfq have been conducted in the absence of ATP. However, destabilization of the Hfq–RNA complexes in the presence of ATP indicates that at least some of the Hfq RNA-binding sites, in the presence of ATP, may allow RNA translocation. Even if Hfq possesses multiple RNA-binding sites in addition to those reported by the Brennan group (Schumacher et al. 2002; Mikulecky et al. 2004; Sun and Wartell 2006), making the final pronouncement on where exactly on Hfq (rA)n may bind seem premature, it seems reasonable to reserve final judgment until the X-ray structure of an Hfq–(rA)n complex is reported and clear evidence indicating that the complex in question is stable in the presence of “physiological” concentrations of ATP is presented. Our finding that ATP nearly abolishes the Hfq footprint at the junction of the model RNA's rA18-tail and the adjacent hairpin and other results suggesting destabilization of Hfq–RNA complexes in the presence of ATP should be taken into account when speculating regarding plausible Hfq–(rA)n interactions in vivo. Even though our fluorescence experiments indicated that competition between ATP and rA7 is unlikely (since controls showed no changes in fluorescence anisotropy in Hfq–RNA complexes in the presence of excess AMP-PNP comparable to those seen with ATP), at present we cannot entirely rule out this possibility. In theory, such competition (modulated by changes in the physiological ATP levels) could regulate the availability of mRNA termini to translation and end modifications via locking and unlocking of the Hfq-bound RNA's (rA)-rich tracts. At present, however, this model is speculative. One of the issues that remains to be clarified in the future is the significance of the Hfq-associated ATPase activity. Whether it is only a latent, unproductive ATP hydrolysis (an “undesirable” consequence of ATP binding) or whether the ATP hydrolysis is coupled in vivo to some as-yet-undetermined form of RNA remodeling remains to be established. Currently we are conducting experiments to clarify these issues.
In summary, ATP binding by Hfq—a central issue of this study—was extensively proven by multiple, independent lines of evidence here, which include (1) binding assays with fluorophore-modified nucleotides; (2) evidence of latent ATP hydrolysis by purified Hfq; (3) EMSA; and (4) RNase A footprinting experiments with unmodified RNAs, which also demonstrate the destabilizing effect of ATP on Hfq–RNA complexes. (5) Hfq–ATP interaction is also indirectly supported by a significant (>50%) homology between the aligned segments representing Hfq's putative ATP-binding site and those in another well-documented ATP-binding enzyme, ClpB, which includes a Walker A-box. (6) Our modeling work further demonstrates compatibility of the predicted ATP-binding cleft with the structure and configuration of an ATP molecule. (7) Finally, we show that knockout of Tyr25—the amino acid forming one of the walls of the predicted ATP-binding cleft—results in reduced ATP binding and reduced ATP hydrolysis in the purified protein. Contrary to a widely circulating model of Hfq as a maverick regulatory protein, the summary of the existing data on Hfq—particularly its physical and functional interaction with the ribosomal protein S1 (Sukhodolets and Garges 2003)—indicates that Hfq may function as a subunit of the translational apparatus. In agreement with this, studies focused on the global effects of Hfq on translation and the stability of multiple RNA species have consistently reported its effects on multiple RNA targets (e.g., see Sonnleitner et al. 2006). It is possible that, by default (when the ATP level in the bacterial cell is reasonably high), mRNAs are continuously threaded through the Hfq pore. This arrangement is insinuated by several indirect lines of evidence. In the only existing high-resolution structure of an Hfq–RNA complex (Schumacher et al. 2002), the 3′-terminus of the Hfq-bound oligonucleotide is located in the Hfq pore. Furthermore, a hypothetical role for Hfq as an mRNA “sliding clamp” is consistent with its known ability to mediate the processivity of RNA-modifying enzymes such as the 3′ → 5′ ribonucleases and the Poly(A) polymerase (Le Derout et al. 2003; Mohanty et al. 2004). It seems likely that further development of this work should clarify the functional significance of utilization of ATP by Hfq–RNA complexes.
Materials and Methods
Proteins
Recombinant Hfq was isolated as previously described (Sukhodolets and Garges 2003), with the sole exception that a Superdex 200HR 10 × 300 column (Amersham Biosciences) was used during the final, gel-filtration stage of the purification procedure. Typically, 450–500 mL of E. coli cultures (induced at an OD600 of 0.6–0.8 with 1 mM IPTG and harvested after 4 h of shaking at 37°C), carrying the plasmids indicated below, were used in each individual purification procedure. This purification procedure yields essentially homogeneous Hfq, with no detectable impurities in the final protein preparations, as judged by silver-stained SDS-gels (Sukhodolets and Garges 2003).
Site-directed mutagenesis
Hfq mutagenesis was performed using the QuikChange site- directed mutagenesis kit (Stratagene) in accordance with the manufacturer's instructions with the wild-type hfq clone in pQE31 Hfq (Sukhodolets and Garges 2003) as a DNA template. To construct the hfq His57Ala mutation, the mutagenic primers MS274 (5′-GAGATGGTTTACAAGGCCGCGATTTCTACTGTT) and MS275 (antiparallel to MS274) were used. To construct the hfq Tyr25Ala mutation, the mutagenic primers MS272 (5′-GTTCCAGTTTCTATTGCTTTGGTGAATGGTATT) and MS273 (antiparallel to MS272) were used. To construct the hfq Gln13Ala mutation, the mutagenic primers MS119 (5′-GATCCGTTCCTGGCCGCACTGCGTCGG) and MS120 (antiparallel to MS119) were used. The entire hfq sequence containing the desired mutations in the resulting plasmids (pQE31 HfqH57A, pQE31 HfqY25A, and pQE31 HfqN13A, respectively) were confirmed by DNA sequencing; two or more clones were selected for each of the mutations. The mutant proteins were overexpressed in M15(pRep4) cells (QIAGEN) and purified as described above.
ATPase activity assays
The ATPase activity assays were, in general, as previously described (Sukhodolets and Garges 2003). Because the ATPase near-knockout Hfq protein described in this study showed particularly low specific ATPase activity, the initial velocities of ATP hydrolysis at relatively high ATP concentrations could not be determined accurately, which hindered the determination of K m and V max for the mutant protein. Therefore, specific activities of the wild-type and mutant proteins referred to in ▶ were measured at >2 K m,ATP values under the previously described conditions (in 50 mM Tris-HCl at pH 7.5, 100 mM NaCl, and 2 mM MgCl2 at 37°C; the typical incubation times were 30–90 min).
Electron microscopy and image analysis
Aliquots of Hfq (in the presence or absence of the ligands indicated below) were adsorbed to 300 mesh carbon-coated grids and stained with 1% uranyl acetate. The following conditions were tested: Hfq alone (either wild-type Hfq or the Hfq Y25A mutant), complexed to rA7 oligonucleotide (10 μM), to ATP (1 mM), to rA7 oligonucleotide and ATP (10 μM and 1 mM, respectively), to rA7 oligonucleotide and ATP-γ-S (10 μM and 1 mM), and to rA7 oligonucleotide and AMP-PNP (10 μM and 1 mM). Samples were viewed using a Philips CM120 electron microscope at an accelerating voltage of 120 kV and nominal magnification of 75,000×. Images having a final pixel size of 0.26 nm were recorded using a 1k GATAN ssCCD camera. For each condition, a total of ∼3000 single particles were windowed and aligned by using X-MIPP software (Marabini 1996 #109) before classification by a self-organizing neural network (Marabini 1994 #110). Rotational power spectra analysis was performed from each average image of the different protein samples.
Fluorescence anisotropy measurements
Fluorescence anisotropy measurements were carried out using a PTI A1010 fluorimeter. In order to determine the Hfq affinity for RNA oligonucleotides, samples containing 5′-fluoresceinated oligonucleotides (Dharamacon) were excited at 490 nm, and emission was measured at 520 nm. The binding buffer contained 20 mM Tris-HCl (pH 8.0) and 100 mM NaCl (Buffer A). One nanomolar 5′-fluoresceinated oligonucleotide was titrated with Hfq, using a 1-mL cuvette; the reactions were incubated for 5 min before the measurements were taken. The molar Hfq concentrations refer to the moles of Hfq hexamer. The measurements were carried out at 298°K. The normalization of fluorescence anisotropy was carried out after determination of the ΔAmax value, which was obtained at saturating Hfq concentrations. ΔA/ΔAmax ratios were determined and plotted versus the Hfq concentrations. The data were plotted using Sigmaplot software, and the curves were fit by the nonlinear least-squares regression method, assuming a bimolecular model: Hfq(hexamer) + rN7 ↔ Hfq(hexamer):rN7 (thus assuming a 1:1 Hfq:RNA stoichiometry) based on the reported Hfq:rN7 crystal structure (Schumacher et al. 2002).
In order to determine the number of Hfq ATP-binding sites in the Hfq hexamer, stoichiometric titrations were carried out. Samples containing MANT nucleotides [3′(2′)-O-(N-methylantraniloyl nucleotide; Jenabioscience] were excited at 355 nm, and emission was measured at 448 nm. The measurements were performed at 298°K. The binding buffer was Buffer A supplemented with 2 mM MnCl2. Typically, 10 μM MANT nucleotide was titrated with Hfq in 1-mL cuvettes. Affinity for MANT nucleotides was determined in a similar fashion, with the exception that MANT-ATP, MANT-ADP, or MANT-AMP-PNP was used at 50 nM. In both types of assays, the molar Hfq concentrations refer to the moles of Hfq monomer. The data were plotted using Kaleidagraph, and during the affinity measurements, the curves were fit using a nonlinear least-squares regression method, assuming the following model: Hfq(monomer) + A ↔ Hfq:A. Control experiments indicated that unmodified nucleotides compete with MANT-ATP for binding to Hfq. At 1 μM MANT-ATP, in the presence of Hfq, addition of 1 μM of the (unlabeled) ATP resulted in approximately twofold reduction of the gained fluorescence anisotropy; addition of 5 μM of unlabeled ATP to 1 μM MANT-ATP in the presence of Hfq was accompanied by an approximately fourfold reduction in fluorescence anisotropy, indicating a competition between labeled and unlabeled nucleotides. Similar controls indicated a competition between labeled and unlabeled RNA oligonucleotides (data not shown).
Coupled in vitro transcription–translation experiments
Coupled in vitro transcription–translation assays were carried out with S30 whole-cell extracts prepared from the hfq− (MC4100) E. coli strain as previously described (Sukhodolets and Garges 2003). The reactions and the immunodetection of the luciferase by Western blotting were essentially as described (Sukhodolets and Garges 2003).
RNA footprinting
The RNA probe (5′-CCUGGUUUUUAAGGAGUGUCGCCAGAGUGCCGCGAUGAAAAAAAAAAAAAAAAAA; Dharmacon) incorporating a stem–loop structure followed by an rA tail was labeled at the 5′-end using T4 polynucleotide kinase (USB) and [γ-32P]ATP, according to the USB protocol. Following the end-labeling procedure, the RNA was gel-purified on a denaturing 20% polyacrylamide gel (National Diagnostics). The gel-purification procedure was as follows. After PAGE, the “wet” gel was covered with a plastic wrap and exposed to an X-ray film. The visualized full-length RNA band was marked and cut from the gel. The polyacrylamide gel slice containing end-labeled RNA was then manually homogenized in an RNase-free, 1.5-mL Eppendorf-tube-size disposable homogenizer, typically, using 200 μL of 2× TBE supplemented with 1 μL of ribonuclease inhibitor (Invitrogen, 10 U/μL). Following the 2–3-min homogenization, the slurry was immediately applied on a microcentrifuge filter vial (Ultrafree-DA; Millipore), and the flow-thru was aliquoted and stored at −70°C. Ribonuclease A footprinting reactions were carried out in 50 mM Tris-HCl (pH 7.9), 10 mM MgCl2, and 1 mM dithiothreitol (Buffer B) containing 50, 100, or 200 mM NaCl. Five-microliter footprinting reactions included 0.5 μL of 10× Buffer B plus NaCl, 0.5 μL of 10 mM ATP or ATP-γ-S (if present), 0.2–1.6 μL of Hfq (typically, 1 mg/mL Hfq glycerol stocks, which were stored at −20°C, were diluted three- to fivefold with purified, nuclease-free water), 0.5 μL of end-labeled RNA probe (4000–6000 cpm/μL), and purified, nuclease-free water to a final volume of 5 μL. After 5 min at room temperature, 1 μL of Ribonuclease A (Boehringer DNase-free RNase; the 2 U/μL stock solutions were diluted to 0.0002 U/μL) was added, and following a 30-sec digestion, 1 μL of 1% SDS was added to stop the reactions. Six microliters of Loading Buffer (0.1% Bromophenol Blue, 40% glycerol, 100 mM EDTA at pH 8) was added to each reaction, and 4–6-μL aliquots of the samples were analyzed on denaturing 20% polyacrylamide gels, which were cast using the Sequagel Sequencing System Kit (National Diagnostics). Model S2 “sequencing” gel apparatuses (BRL) were used; runs were carried out at 50 W. Following the electrophoresis, the “wet” gels were transferred onto a film support and covered with a plastic wrap, and F-BX810 X-ray films (Phenix Research Products) were exposed to gels at −70°C with BioMax MS Screens (Kodak).
EMSA
EMSA experiments were carried as described (Sukhodolets et al. 2006) with the exception that 0.25-mm-thick 14% polyacrylamide gels containing 6 M urea (which were cast using the Sequagel Kit; National Diagnostics) were used to monitor the formation of high-affinity (urea-insensitive) Hfq–RNA complexes.
Model S2 “sequencing” gel apparatuses (BRL) were used in these experiments. Typically, binding reactions included 0.2 μM Hfq (in Buffer B containing 100 mM NaCl) and, if present, 1 mM ATP.
A structural model for the Hfq–ATP complex
A structural model for the Hfq–ATP complex was built by initial positioning of the adenine moiety parallel and ∼3.4 Å above the plane of the Y25 phenol side chain, followed by manipulation of the ATP dihedral angles; both glycosidic and phosphoester backbone torsion angles were adjusted so as to optimize favorable interactions (electrostatic and hydrogen bonding) with the local protein environment. The PyMOL software package and its Python application programming interface were used for modeling, docking, and visualization tasks, as well as all molecular graphics illustrations. A Python module was written to use primitives from PyMOL's Compiled Graphics Objects facility in conjunction with the geometric properties of distance-weighted adjacency matrices in order to enable the production of polygonal, plate-like representations of cyclic molecular structures (e.g., the Y25 and ATP rings); the module is available from the authors upon request.
Acknowledgments
We thank P. England and colleagues at the PBFMI/Paris Pasteur Institute for their help in performing the fluorescence measurements. M.V.S. is indebted to Sankar Adhya for support and help in establishing an independent research laboratory at Lamar University; this transition was also greatly enhanced by a generous gift of equipment and materials from NIH. We thank Byongkook Lee and Bangalore K. Sathyanarayana for stimulating discussions of possible alternative configurations of the Hfq–ATP complex; and Sankar Adhya, Susan Garges, and Karen E. Sukhodolets for discussion of the experimental results and critical comments on the manuscript. V.A. thanks P. Regnier for his interest in this work and Olivier Pellegrini for technical assistance. C.M. thanks J. Andrew McCammon (UCSD) for funding and computational facilities, as well as a Sloan/DOE postdoctoral fellowship. This work was supported in part by a Welch Foundation grant (V-0004).
Footnotes
Reprint requests to: Maxim V. Sukhodolets, Laboratory of Biochemistry, Department of Chemistry, Lamar University, Beaumont, TX 77710, USA; e-mail: msoukhodol@my.lamar.edu; fax: (409) 880-8270.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072883707.
References
- Arluison V., Derreumaux, P., Allemand, F., Folichon, M., Hajnsdorf, E., and Régnier, P. 2002. Structural modeling of the Sm-like protein Hfq from Escherichia coli . J. Mol. Biol. 320 705–712. [DOI] [PubMed] [Google Scholar]
- Arluison V., Mura, C., Guzman, M.R., Liquier, J., Pellegrini, O., Gingery, M., Regnier, P., and Marco, S. 2006. Three-dimensional structures of fibrillar Sm proteins: Hfq and other Sm-like proteins. J. Mol. Biol. 356 86–96. [DOI] [PubMed] [Google Scholar]
- Azam T.A., Hiraga, S., and Ishihama, A. 2000. Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells 5 613–626. [DOI] [PubMed] [Google Scholar]
- Brescia C.C., Mikulecky, P.J., Feig, A.L., and Sledjeski, D.D. 2003. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA 9 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBow M.S., Ryan, T., Young, R.A., and Blumenthal, T. 1977. Host factor for coliphage Q β RNA replication: Presence in procaryotes and association with the 30S ribosomal subunit in Escherichia coli . Mol. Gen. Genet. 153 39–43. [DOI] [PubMed] [Google Scholar]
- Folichon M., Arluison, V., Pellegrini, O., Huntzinger, E., Regnier, P., and Hajnsdorf, E. 2003. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 31 7302–7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franze de Fernandez M.T., Hayward, W.S., and August, J.T. 1972. Bacterial proteins required for replication of phage Q ribonucleic acid. Purification and properties of host factor I, a ribonucleic acid-binding protein. J. Biol. Chem. 247 824–831. [PubMed] [Google Scholar]
- Kajitani M., Kato, A., Wada, A., Inokuchi, Y., and Ishihama, A. 1994. Regulation of the Escherichia coli hfq gene encoding the host factor for phage Q β. J. Bacteriol. 176 531–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Derout J., Folichon, M., Briani, F., Deho, G., Regnier, P., and Hajnsdorf, E. 2003. Hfq affects the length and the frequency of short oligo(A) tails at the 3′ end of Escherichia coli rpsO mRNAs. Nucleic Acids Res. 31 4017–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz D.H., Mok, K.C., Lilley, B.N., Kulkarni, R.V., Wingreen, N.S., and Bassler, B.L. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae . Cell 118 69–82. [DOI] [PubMed] [Google Scholar]
- Marabini R. and Carazo, J.M. 1994. Pattern recognition and classification of images of biological macromolecules using artificial neural networks. Biophys. J. 66 1804–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabini R., Masegosa, I.M., San Martin, C., Marco, S., Fernandez, J.J., de la Fraga, L.G., Vaquerizo, C., and Carazo, J.M. 1996. Xmipp: An image processing package for electron microscopy. J. Struct. Biol. 116 237–240. [DOI] [PubMed] [Google Scholar]
- Masse E., Majdalani, N., and Gottesman, S. 2003. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 6 120–124. [DOI] [PubMed] [Google Scholar]
- Mikulecky P.J., Kaw, M.K., Brescia, C.C., Takach, J.C., Sledjeski, D.D., and Feig, A.L. 2004. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 11 1206–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty B.K., Maples, V.F., and Kushner, S.R. 2004. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli . Mol. Microbiol. 54 905–920. [DOI] [PubMed] [Google Scholar]
- Moll I., Afonyushkin, T., Vytvytska, O., Kaberdin, V.R., and Blasi, U. 2003. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 9 1308–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller T., Franch, T., Hojrup, P., Keene, D., Bächinger, H.P., Brennan, R.G., and Valentin-Hansen, P. 2002. HFq: A bacterial Sm-like protein that mediates RNA–RNA interaction. Mol. Cell 9 23–30. [DOI] [PubMed] [Google Scholar]
- Muffler A., Traulsen, D.D., Fischer, D., Lange, R., and Hengge-Aronis, R. 1997. The RNA-binding protein HF-1 plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the σS subunit of RNA polymerase in Escherichia coli . J. Bacteriol. 179 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura C., Phillips, M., Kozhukhovsky, A., and Eisenberg, D. 2003. Structure and assembly of an augmented Sm-like archaeal protein 14-mer. Proc. Natl. Acad. Sci. 100 4539–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikulin A., Stolboushkina, E., Perederina, A., Vassilieva, I., Blaesi, U., Moll, I., Kachalova, G., Yokoyama, S., Vassylyev, D., Garber, M., et al. 2005. Structure of Pseudomonas aeruginosa Hfq protein. Acta Crystallogr. D Biol. Crystallogr. 61 141–146. [DOI] [PubMed] [Google Scholar]
- Pannone B.K. and Wolin, S.L. 2000. Sm-like proteins wRING the neck of mRNA. Curr. Biol. 10 478–481. [DOI] [PubMed] [Google Scholar]
- Schumacher M.A., Rearson, R.F., Moller, T., Valentin-Hansen, P., and Brennan, R.G. 2002. Structures of the pleiotropic translational regulator HFq and an HFq-RNA complex: A bacterial Sm-like protein. EMBO J. 21 3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta J., Agrawal, R.K., and Frank, J. 2001. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc. Natl. Acad. Sci. 98 11991–11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski D.D., Whitman, C., and Zhang, A. 2001. HFq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 183 1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner E., Schuster, M., Sorger-Domenigg, T., Greenberg, E.P., and Blasi, U. 2006. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa . Mol. Microbiol. 59 1542–1558. [DOI] [PubMed] [Google Scholar]
- Sukhodolets M.V. and Garges, S. 2003. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry 42 8022–8034. [DOI] [PubMed] [Google Scholar]
- Sukhodolets M.V., Garges, S., and Adhya, S. 2006. Ribosomal protein S1 promotes transcriptional cycling. RNA 12 1505–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. and Wartell, R.M. 2006. Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites. Biochemistry 45 4875–4887. [DOI] [PubMed] [Google Scholar]
- Sun X., Zhulin, I., and Wartell, R.M. 2002. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 30 3662–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui H.C.T., Feng, G., and Winkler, M.E. 1996. Transcription of the mutL repair, miaA tRNA modification, hfq pleiotropic regulator, and hflA region protease genes of Escherichia coli K12 from clustered Es32-specific promoters during heat-shock. J. Bacteriol. 178 5719–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui H.C.T., Feng, G., and Winkler, M.E. 1997. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 179 7476–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui H.C.T., Leung, H.C.E., and Winkler, M.E. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 13 35–49. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P., Eriksen, M., and Udesen, C. 2004. The bacterial Sm-like protein Hfq: A key player in RNA transactions. Mol. Microbiol. 51 1525–1533. [DOI] [PubMed] [Google Scholar]
- Vytvytska O., Jakobsen, J.S., Balcunate, G., Andersen, J.S., Baccarini, M., and von Gabain, A. 1998. Host-factor I, Hfq, binds to Escherichia coli ompA mRNA in a growth rate-dependent fashion and regulates its stability. Proc. Natl. Acad. Sci. 95 14118–14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman K.M. 2002. Small RNAs in bacteria: Diverse regulators of gene expression in response to environmental changes. Cell 109 141–144. [DOI] [PubMed] [Google Scholar]
- Zhang D., Abovich, N., and Rosbash, M. 2001. A biochemical function for the Sm complex. Mol. Cell 7 319–329. [DOI] [PubMed] [Google Scholar]
- Zhang A., Wassarman, K.M., Ortega, J., Steven, A.C., and Storz, G. 2002. The Sm-like HFq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9 11–22. [DOI] [PubMed] [Google Scholar]
- Zylicz M., LeBowitz, J.H., McMacken, R., and Georgopoulos, C. 1983. The dnaK protein of Escherichia coli possesses an ATPase and autophosphorylating activity and is essential in an in vitro DNA replication system. Proc. Natl. Acad. Sci. 80 6431–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]