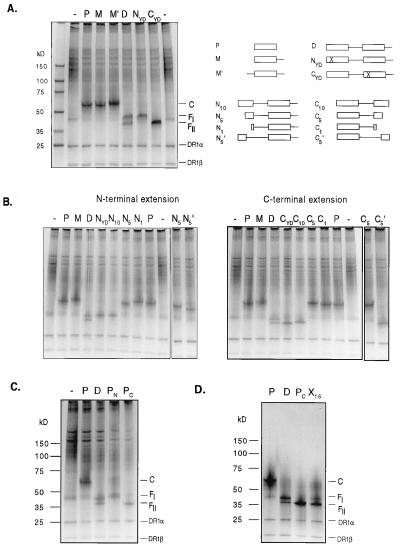
Figure 2.
Induction of FI- and FII-complexes by amino acid overhangs of the ligand. Soluble HLA-DR1 molecules were loaded with ligand constructs containing the epitope HA306-318. The structure of the ligands is depicted schematically on the upper right panel. The box represents the HA306-318 binding epitope, and the line represents a tetraethylene glycol unit used to connect the epitopes. The cross in the NYD- and the CYD-ligand marks a substitution of the main anchor residue Y308 with D in one of the two HA306-318 epitopes, which abrogates binding to the HLA-DR1 molecule. N10-N1 and C10-C1 represent dimeric ligand constructs with truncations in the N- or C-terminal epitope. (A) Formation of FI- and FII-ligand complexes with teg-linked peptide constructs with defined N- or C-terminal polypeptide extensions. HLA-DR1/ligand complexes, loaded with monomeric (lanes P, M, and M′) and dimeric HA306-318 ligand constructs (lanes D, NYD, and CYD), were separated by SDS/PAGE. The position of bands representing the compact C-form (associated with the monomeric ligands) as well as of the two distinct FI- and FII-forms (associated with the dimeric ligands) are indicated. (B) To estimate distance and size of the polypeptide region responsible for the formation of the FI- and FII-complexes, HLA-DR1 molecules were loaded with truncated forms of the dimeric HA306-318 construct. The series of N-terminal truncations (Left) consists of HA306-318 peptides linked by a teg spacer with the extension VKQNTLKLAT (lane N10), LKLAT (lane N5), and T (lane N1). In addition, a ligand also was used in which the LKLAT overhang was connected by two instead of one teg-units (lane N5′, shown on the right side of the gel). For the series of C-terminal truncations (Right), overhangs were used with the sequence GPKYVKQNTL (lane C10), GPKYV (lanes C5 and C5′), and G (lane C1). On both gels, empty HLA-DR1 molecules (−) and ligand complexes preformed with the HA306-318 peptide (P lanes), the monomer (M lanes), and the dimer (D lanes) are shown as a reference. (C) Formation of F-complexes with naturally extended polypeptide ligands. HLA-DR1 molecules were loaded with peptides extended by 20 amino acids of the natural sequence of the hemagglutinin protein on the N-terminal side (HA286-318; lane PN) and on the C-terminal side (HA306-338; lane PC) of the HA binding epitope. SDS/PAGE separation is shown in comparison to the HA306-318-loaded complex (lane P) and the complex loaded with the dimer (lane D). (D) Formation of an FII-complex by a ligand with randomized overhang. HLA-DR1 molecules were loaded with an HA306-318 epitope, which (through a teg-spacer on the C-terminal side) was connected to a 15-aa-long peptide with random sequence (HA-X15). The overhang was produced by a sequential synthesis using an amino acid pool (G, A, P, V, I, L, M, Y, F, S, T, N, Q, D, E, K, R, H). The HLA-DR1/HA-X15 ligand complex (lane X15) is shown in comparison to the complex loaded with the HA306-318-peptide (lane P), the dimer (lane D), and the HA306-338 peptide (lane PC).