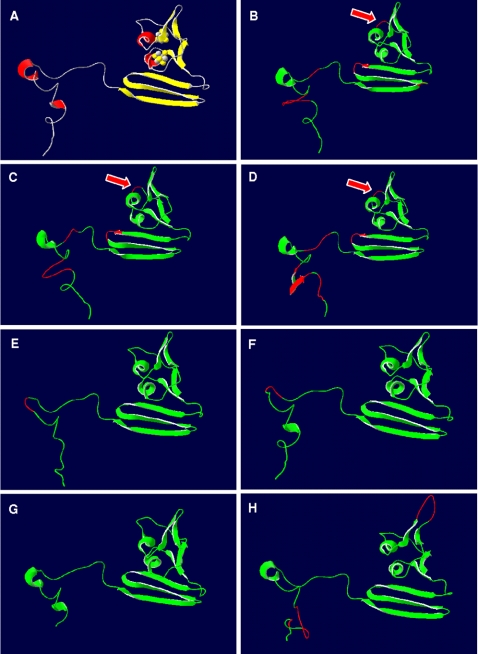
Figure 3. Three-dimensional structure of AprB from A. fulgidus (A) and selected, homology modeling-based AprB models from Allochromatium vinosum (B) and Pelagibacter ubique (C) (as representatives of SOB from Apr lineage-I), Pyrobaculum calidifontis (D) (as representative of crenarchaeal SRP), Desulfotomaculum reducens (E) (as representative of Gram-positive SRB and LGT-affected deltaproteobacterial SRB), Desulfovibrio vulgaris (F) (as representative of non-LGT-affected deltaproteobacterial SRB), Chlorobaculum tepidum (G) and Thiobacillus denitrificans (H) (as representatives of LGT-affected SOB from Apr lineage-II).
Ribbon structure shown from front view (positions of [4Fe-4S] clusters indicated in A. fulgidus AprB). Ribbon structure of A. fulgidus AprB (A) colored by secondary structure elements; ribbon structures of AprB models (B–H) colored by model confidence factor provided by SWISS-MODEL (green, respective region of model and reference structure superpose; red, respective region of model deviates from the reference structure). The missing flexible loop between Cys-B13 and Gly-B19 (enumeration based on A. fulgidus sequence) in models of SOB from Apr lineage-I and Pyrobaculum spp. is marked by red arrows.