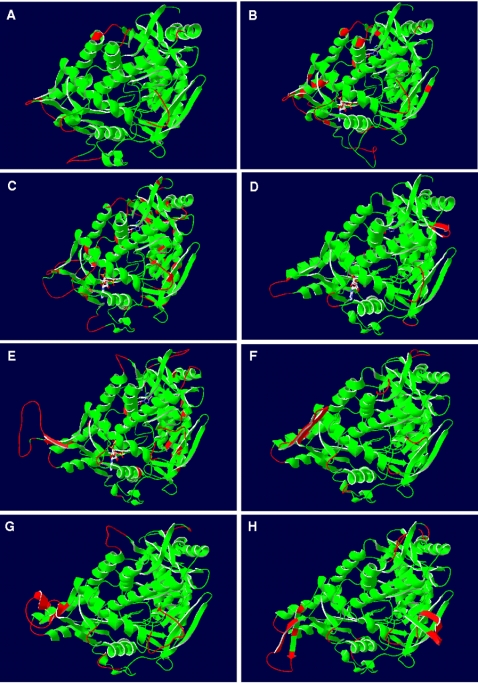
Figure 7. Selected, homology modeling-based AprA models from Allochromatium vinosum (A) and Pelagibacter ubique (B) (as representatives of SOB from Apr lineage-I), Pyrobaculum calidifontis (C) (as representative of crenarchaeal SRP), Desulfotomaculum reducens (D) (as representative of Gram-positive SRB and LGT-affected deltaproteobacterial SRB), Thermodesulfobacterium commune (E) (as representative of thermophilic SRB), Desulfovibrio vulgaris (F) (as representative of non-LGT-affected deltaproteobacterial SRB), Chlorobaculum tepidum (G) and Thiobacillus denitrificans (H) (as representatives of LGT-affected SOB from Apr lineage-II).
Ribbon structure shown from front view (position of FAD cofactor and substrate APS are indicated). Ribbon structure of AprA models colored by model confidence factor provided by SWISS-MODEL (green, respective region of model and reference structure superpose; red, respective region of model deviates from the reference structure).