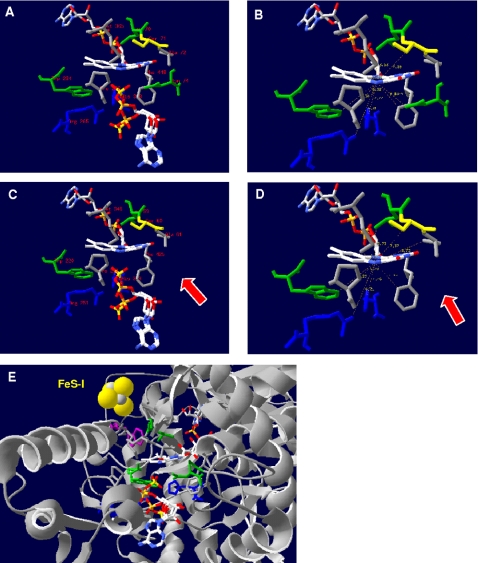
Figure 9. AprA active center from the protein of A. fulgidus (A, B) and the homology modeling-based model of Pyrobaculum calidifontis (C, D) (residues in a distance of less than 6.5 Å to the N5 atom of FAD cofactor are shown).
FAD cofactor and substrate APS (in two conformations) are shown as ball-and-stick representations. Residues involved in the isoalloxazine binding (e.g. A. fulgidus: Leu-A70, Asn-A74, Trp-A234) are highlighted by green color (missing Asn-A63 in the Pyrobaculum calidifontis AprA model is marked by an arrow), the invariant, positively charged residues His-A398 and Arg-A265 of the active site are blue colored (other AA are colored in grey). Distances are given in Å (B, D). (E) The position of the electron-transferring [4Fe-4S] cluster I and Trp-B48 (highlighted by violet color) of AprB to the FAD cofactor in the AprA protein of A. fulgidus is shown (ribbon structure is colored in grey).