Abstract
We have developed and validated a consolidated bead-based genotyping platform, the Bioplex suspension array for simultaneous detection of multiple single nucleotide polymorphisms (SNPs) of the ATP-binding cassette transporters. Genetic polymorphisms have been known to influence therapeutic response and risk of disease pathologies. Genetic screening for therapeutic and diagnostic applications thus holds great promise in clinical management. The allele-specific primer extension (ASPE) reaction was used to assay 22 multiplexed SNPs for eight subjects. Comparison of the microsphere-based ASPE assay results to sequencing results showed complete concordance in genotype assignments. The Bioplex suspension array thus proves to be a reliable, cost-effective and high-throughput technological platform for genotyping. It can be easily adapted to customized SNP panels for specific applications involving large-scale mutation screening of clinically relevant markers.
Keywords: Genotype; Microspheres; Polymorphism, Genetic
Introduction
The advances in biomedical science have led to the discovery of genes that may influence therapeutic outcome and disease susceptibility. Inter-individual variability in drug response can often be attributed to genetic polymorphisms in genes encoding drug metabolizing enzymes, transporters, ion channels and drug target receptors (1-4). Genetic screening for therapeutic and diagnostic applications thus holds great promise though poorly realized at the present moment, owing largely to technological limitations. The factors that hinder pharmacogenetic testing and molecular diagnosis include the laborious and time-consuming nature of genotyping, the fidelity of the genotyping results, as well as the cost incurred by the test (5). Therefore, there is a need to develop a reliable, cost-effective and high-throughput technological platform for genotyping markers involved in the aetiology of variable response to drug therapy or disease pathologies. The Bioplex suspension array is a potential candidate for the development of such a genotyping tool.
The Luminex microbead system has recently been used to define single nucleotide polymorphisms (SNPs) of human leukocyte antigens (6, 7), minor histocompatibility antigens (8), drug metabolizing genes (9, 10), disease genes (11-14), or detect microorganisms (15-18). Despite this, the use of the bead technology as a genotyping tool is currently still pretty much in infancy. Luminex/BioRad offers 100 distinct sets of color-coded tiny beads, called microspheres. The polystyrene beads are internally-dyed with differing ratios of two spectrally distinct fluorophores. The xMAP technology is a simultaneous multiplex bioassay using a rapid flow cytometer that permits detection of multiple analyses bound to the microbeads in solution of a single tube (19, 20). We describe a genotyping assay that combines an allele-specific primer extension (ASPE) with the Bioplex suspension array. The assay relies on the sequence-specific primer extension of two allele-specific capture oligonucleotide probes that differ at the last base of the 3’-end nucleotide defining the alleles. A tag sequence at the 5’-end of the capture probe allows the resulting enzymatic ASPE product to be captured by its complementary sequence (Anti-tags) which has been coupled to a specific fluorescent microsphere (Fig. 1).
Fig. 1.
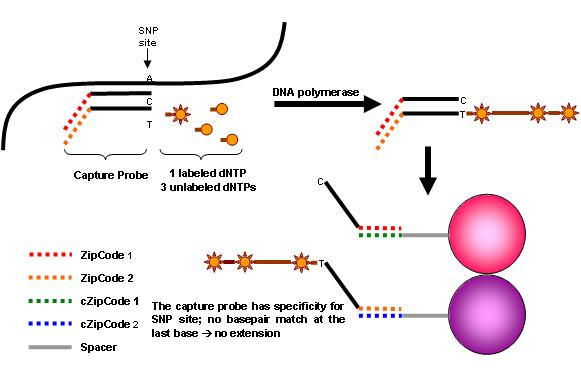
Schematic diagram of ASPE reaction.
This study attempts to exploit the bead-based technological platform to rapidly screen for a basket of genetic markers that may impact on drug response. We have thus developed and validated a multiplex genotyping platform using the Bioplex suspension array for detecting a panel of ATP binding cassette (ABC) drug transporter polymorphisms. Here, we demonstrate the ability of the Bioplex suspension array to simultaneously genotype 22 SNPs in a single reaction (Table 1), with minimal cross-reactivity. This report provides detailed information including primer sequences, anti-tag/microsphere combinations and experimental conditions, hence enabling easy adaptation of the assay to customized SNP panels. This thus represents an important milestone in the development of genotyping technologies and a major step forward in ushering the realization of personalized medicine, as well as the use of molecular diagnosis in clinical management.
Table 1.
Coupling of anti-tags to their corresponding Bioplex beads.
| SNP ID | Bead No. | |
| WT | mt | |
| ABCB1_rs1128503 | 24 | 66 |
| ABCB1_rs2032582 | 33 | 75 |
| ABCB1_rs1045642 | 17 | 61 |
| ABCC1_rs504348 | 19 | 51 |
| ABCC5_ rs1000002 | 21 | 53 |
| ABCC5_ rs3749438 | 27 | 54 |
| ABCC5_ rs4148557 | 35 | 56 |
| ABCC5_rs562 | 37 | 62 |
| ABCC5_rs1132776 | 42 | 64 |
| ABCC5_i1-1205 | 43 | 72 |
| ABCC5_i1-1679 | 45 | 74 |
| ABCC5_i1-1821 | 47 | 77 |
| ABCC4_rs1479390 | 6 | 34 |
| ABCC4_rs2274405 | 11 | 36 |
| ABCC4_rs1189429 | 12 | 38 |
| ABCC4_rs4148436 | 13 | 41 |
| ABCC4_ rs1189437 | 14 | 44 |
| ABCC4_ rs1151471 | 18 | 46 |
| ABCC4_ rs1059762 | 20 | 52 |
| ABCC4_rs868853 | 25 | 55 |
| ABCC4_rs9524885 | 26 | 63 |
| ABCC4_rs869951 | 28 | 65 |
Materials and Methods
Oligonucleotides
All oligonucleotides, unless otherwise stated, were synthesized by Research Biolabs, Singapore. The PCR primers used for generating the various fragments encompassing the polymorphism sites (Table 2) were unmodified and purified by standard desalting procedures. A 24-mer universal tag sequence was incorporated at the 5’-end of each ASPE sequence. The ASPE sequences (18-24mer) were designed to be matched for melting temperature at 51-56°C. The tag sequences were designed using the web-based ‘Tag-IT’ Oligo Design software (Tm Bioscience Corporation, Toronto, Ontario, Canada). The TAG-ASPE primers were unmodified and HPLC-purified (Table 3). The complementary oligos (Anti-tags) were amino (NH2)-modified at 5’-end for covalent attachment to carboxylated microspheres and separated from the anti-tag sequences (Table 4) by a 6-carbon spacer to minimize potential interactions with the micropsheres. Anti-tags were purified by standard desalting procedures and obtained from Sigma-Proligo, Singapore.
Table 2.
PCR primer sequences and amplification conditions.
The annealing temperatures of the various multiplex PCR panels are indicated in parentheses.
| Panel A (54°C) | Primer concentration (µM) | PCR Product Size (bp) |
| ABCB1_rs1128503F: TCTTTGTCACTTTATCCAGC | 0.08 | 502 |
| ABCB1_rs1128503R: TCTCACCATCCCCTCTGT | 0.08 | |
| ABCB1_ rs2032582F: TGCAGGCTATAGGTTCCAGG | 0.06 | 284 |
| ABCB1_ rs2032582R: TAGGGAGTAACAAAATAACAC | 0.06 | |
| ABCB1_rs1045642F: CTCACAGTAACTTGGCAG | 0.14 | 315 |
| ABCB1_rs1045642R: CTTACATTAGGCAGTGAC | 0.14 | |
| ABCC1_rs504348F: CAGGATGAAATGAGGGCACAG | 0.12 | 155 |
| ABCC1_rs504348R: GAAGCGCCTGGGATCTTTGG | 0.12 | |
| Panel B (56°C) | ||
| ABCC5_ rs3749438F: GGGTGAAATGGAACTGACTC | 0.03 | 390 |
| ABCC5_ rs3749438R: GGTGCCCAGGAAACAGAG | 0.03 | |
| ABCC5_i1-1821/1679F: CAACATATATGAAGTATTTCAGCGG | 0.35 | 770 |
| ABCC5_i1-1821/1679R: GAAATATCTTTATGAACTTGGGAG | 0.35 | |
| ABCC5_i1-1205F: CAAGAATATGCTGCTTTACG | 0.16 | 859 |
| ABCC5_i1-1205R: TAACCCGTTGAGAGTCGTCA | 0.16 | |
| ABCC5_rs1132776F: AGCCCAGGGTCATATGAACAGAGA | 0.02 | 477 |
| ABCC5_rs1132776R: AGGCAAAAAGTCAACAACAACCAG | 0.02 | |
| ABCC5_rs562F: GCAGGTCCCAAAGCCATTCAGG | 0.03 | 408 |
| ABCC5_rs562R: ACCGCAGTCGTCGCACAGTCTCT | 0.03 | |
| ABCC5_rs1000002F: CTGGCCTGTCCTAGCTGGGTATG | 0.05 | 291 |
| ABCC5_rs1000002R: CTCTGCCTCTTCCTCTTTGCTTCC | 0.05 | |
| ABCC5_ rs4148557F: GTTGGGGCGGTGAGCAGTTTG | 0.04 | 316 |
| ABCC5_ rs4148557R: TTCCCCACACCCTTCATACATAGA | 0.04 | |
| Panel C (60°C) | ||
| ABCC4_ rs1189429F: AGTGACAGTTATTGAGGTTTC | 0.12 | 290 |
| ABCC4_ rs1189429R: ATCTGCCTCTTTCCACTCC | 0.12 | |
| ABCC4_ rs1189437F: ACACCATCTCTACTAAAAATAC | 0.20 | 457 |
| ABCC4_ rs1189437R: CAGAGACCACACCAACCAC | 0.20 | |
| ABCC4_ rs868853F: CGCCCGACCAATATCTCACTTTT | 0.10 | 230 |
| ABCC4_ rs868853R: TCCTACAGCCATTCAACCAGCATA | 0.10 | |
| ABCC4_rs869951F: GCCTGCGCCGCTGGATGGA | 0.05 | 393 |
| ABCC4_rs869951R: TCGAGTTACCCGGCTTTCTTGAGG | 0.05 | |
| ABCC4_ rs4148436F: GGTGTCTGTGTGTGGGGAGT | 0.04 | 482 |
| ABCC4_ rs4148436R: TAAGAGTGAACCCTGCCACA | 0.04 | |
| ABCC4_ rs1479390F: CACTGCCAGGGCCTCAAAATGTA | 0.07 | 620 |
| ABCC4_ rs1479390R: ATGCAGCTGCCAAATGGAAGTCTA | 0.07 | |
| ABCC4_ rs1151471F: TGACTCTTGGTTCCTCTATAGC | 0.15 | 202 |
| ABCC4_ rs1151471R: AGGACACAATATAACATCTTGC | 0.15 | |
| Panel D (56°C) | ||
| ABCC4_ rs1059762F: CCTCTCAGAATAAGGTGTCAC | 0.10 | 437 |
| ABCC4_ rs1059762R: CATTAAAACAGAAACAGGACG | 0.10 | |
| ABCC4_ rs2274405F: CACAGCCCCATACAGCGTCACT | 0.10 | 318 |
| ABCC4_ rs2274405R: Tttttgttgttgttgcccaggatg | 0.10 | |
| ABCC4_ rs9524885F: AGGAATGGAGGGAATGAGTT | 0.15 | 637 |
| ABCC4_ rs9524885R: CTTGTAGAACGTGATCAAAATG | 0.15 |
Table 3.
TAG-ASPE primer sequences.
| Tagged ABCB1_rs1128503wt: TACATACACTAATAACATACTCAT CTGGTAGATCTTGAAGGGC |
| Tagged ABCB1_rs1128503mt: CAATTTACTCATATACATCACTTT CTGGTAGATCTTGAAGGGT |
| Tagged ABCB1_ rs2032582wt: ATACTACATCATAATCAAACATCA ATAAGAAAGAACTAGAAGGTG |
| Tagged ABCB1_ rs2032582mt: ATCATACATACATACAAATCTACA ATAAGAAAGAACTAGAAGGTT |
| Tagged ABCB1_rs1045642wt: CAATTAACTACATACAATACATAC TGGTGTCACAGGAAGAGATC |
| Tagged ABCB1_rs1045642mt: CTACAAACAAACAAACATTATCAA TGGTGTCACAGGAAGAGATT |
| Tagged ABCC1_rs504348wt: AATCAATCTTCATTCAAATCATCA GGATACTGTCCTTAAACAGC |
| Tagged ABCC1_rs504348mt: TCAATCAATTACTTACTCAAATAC GGATACTGTCCTTAAACAGG |
| Tagged ABCC4_rs2274405wt: CTACTATACATCTTACTATACTTT ATTTTGCTTGCACTGAAAAAT |
| Tagged ABCC4_rs2274405mt: CTATCTATCTAACTATCTATATCA ATTTTGCTTGCACTGAAAAAC |
| Tagged ABCC4_rs1189429wt: CTTTCTATCTTTCTACTCAATAAT ggtgatctcatgcccttg |
| Tagged ABCC4_rs1189429mt: CTTTTACAATACTTCAATACAATC ggtgatctcatgccctta |
| Tagged ABCC4_ rs1189437wt: TCATTCATATACATACCAATTCAT agtattcttttcaaaaatacttg |
| Tagged ABCC4_ rs1189437mt: TCAATTACTTCACTTTAATCCTTT agtattcttttcaaaaatacttt |
| Tagged ABCC4_rs868853wt: CAATTTCATCATTCATTCATTTCA ccattcaaggttatccttac |
| Tagged ABCC4_rs868853mt: TTACTTCACTTTCTATTTACAATC ccattcaaggttatccttat |
| Tagged ABCC4_rs869951wt: TTCAATCATTCAAATCTCAACTTT ttctcaggaccaaacgac |
| Tagged ABCC4_rs869951mt: AAACTAACATCAATACTTACATCA ttctcaggaccaaacgag |
| Tagged ABCC4_rs9524885wt: TCATTTACCAATCTTTCTTTATAC tgctgatgctgctaatcct |
| Tagged ABCC4_rs9524885mt: CTATCTTTAAACTACAAATCTAAC tgctgatgctgctaatccc |
| Tagged ABCC4_rs4148436wt: TCATTTCAATCAATCATCAACAAT agccccatcagtagcaca |
| Tagged ABCC4_rs4148436mt: TCAATTACCTTTTCAATACAATAC agccccatcagtagcacg |
| Tagged ABCC4_rs1479390wt: TACACTTTCTTTCTTTCTTTCTTT aattgttgtgagtccacttg |
| Tagged ABCC4_rs1479390mt: TTACCTTTATACCTTTCTTTTTAC aattgttgtgagtccacttt |
| Tagged ABCC4_rs1151471wt: CAATAAACTATACTTCTTCACTAA aaattgtagctataagatgatc |
| Tagged ABCC4_rs1151471mt: TCATTTCACAATTCAATTACTCAA aaattgtagctataagatgatt |
| Tagged ABCC4_rs1059762wt: AATCTTACTACAAATCCTTTCTTT acattttgaatatagctatcg |
| Tagged ABCC4_rs1059762mt: AATCCTTTTACATTCATTACTTAC acattttgaatatagctatca |
| Tagged ABCC5_ rs1000002wt: CTTTTCATCTTTTCATCTTTCAAT TAGCTCTGATGGTTCTCAC |
| Tagged ABCC5_ rs1000002mt: TTACTTCACTTTCTATTTACAATC TAGCTCTGATGGTTCTCAT |
| Tagged ABCC5_ rs4148557wt: TCATTTACTCAACAATTACAAATC CGGTGAGCAGTTTGAAACA |
| Tagged ABCC5_ rs4148557mt: CTTTAATCTCAATCAATACAAATC CGGTGAGCAGTTTGAAACG |
| Tagged ABCC5_ rs3749438wt: CTTTTCATCAATAATCTTACCTTT GCACTTGGTATGTTCCCG |
| Tagged ABCC5_ rs3749438mt: CTTTAATCCTTTATCACTTTATCA GCACTTGGTATGTTCCCA |
| Tagged ABCC5_i1-1821wt: TCAAAATCTCAAATACTCAAATCA ACACCCAATCTCATTCATAA |
| Tagged ABCC5_i1-1821mt: AATCTAACAAACTCATCTAAATAC ACACCCAATCTCATTCATAG |
| Tagged ABCC5_i1-1679wt: CTAACTAACAATAATCTAACTAAC TGACAAAGTTTTTGAATAATAAA |
| Tagged ABCC5_i1-1679mt: TACACTTTAAACTTACTACACTAA TGACAAAGTTTTTGAATAATAAT |
| Tagged ABCC5_i1-1205wt: TCATCAATCTTTCAATTTACTTAC ACTTGTCCATCTTTATAACAG |
| Tagged ABCC5_i1-1205mt: TCATCAATCAATCTTTTTCACTTT ACTTGTCCATCTTTATAACAA |
| Tagged ABCC5_rs1132776wt: CTATCTTCATATTTCACTATAAAC GATGCTCTGGAAGTACCCA |
| Tagged ABCC5_rs1132776mt: TCATAATCTCAACAATCTTTCTTT GATGCTCTGGAAGTACCCG |
| Tagged ABCC5_rs562wt: TATATACACTTCTCAATAACTAAC GCAACGCTGACCATTCAAT |
| Tagged ABCC5_rs562mt: AATCTACACTAACAATTTCATAAC GCAACGCTGACCATTCAAC |
| The tag sequences are indicated in italics. |
Table 4.
Anti-tag primer sequences.
| Anti-Tagged ABCB1_rs1128503wt: ATGAGTATGTTATTAGTGTATGTA |
| Anti-Tagged ABCB1_rs1128503mt: AAAGTGATGTATATGAGTAAATTG |
| Anti-Tagged ABCB1_ rs2032582wt: TGATGTTTGATTATGATGTAGTAT |
| Anti-Tagged ABCB1_ rs2032582mt: TGTAGATTTGTATGTATGTATGAT |
| Anti-Tagged ABCB1_rs1045642wt: GTATGTATTGTATGTAGTTAATTG |
| Anti-Tagged ABCB1_rs1045642mt: TTGATAATGTTTGTTTGTTTGTAG |
| Anti-Tagged ABCC1_rs504348wt: TGATGATTTGAATGAAGATTGATT |
| Anti-Tagged ABCC1_rs504348mt: GTATTTGAGTAAGTAATTGATTGA |
| Anti-Tagged ABCC4_rs2274405wt: AAAGTATAGTAAGATGTATAGTAG |
| Anti-Tagged ABCC4_rs2274405mt: TGATATAGATAGTTAGATAGATAG |
| Anti-Tagged ABCC4_rs1189429wt: ATTATTGAGTAGAAAGATAGAAAG |
| Anti-Tagged ABCC4_rs1189429mt: GATTGTATTGAAGTATTGTAAAAG |
| Anti-Tagged ABCC4_ rs1189437wt: ATGAATTGGTATGTATATGAATGA |
| Anti-Tagged ABCC4_ rs1189437mt: AAAGGATTAAAGTGAAGTAATTGA |
| Anti-Tagged ABCC4_rs868853wt: TGAAATGAATGAATGATGAAATTG |
| Anti-Tagged ABCC4_rs868853mt: GATTGTAAATAGAAAGTGAAGTAA |
| Anti-Tagged ABCC4_rs869951wt: AAAGTTGAGATTTGAATGATTGAA |
| Anti-Tagged ABCC4_rs869951mt: TGATGTAAGTATTGATGTTAGTTT |
| Anti-Tagged ABCC4_rs9524885wt: GTATAAAGAAAGATTGGTAAATGA |
| Anti-Tagged ABCC4_rs9524885mt: GTTAGATTTGTAGTTTAAAGATAG |
| Anti-Tagged ABCC4_rs4148436wt: ATTGTTGATGATTGATTGAAATGA |
| Anti-Tagged ABCC4_rs4148436mt: GTATTGTATTGAAAAGGTAATTGA |
| Anti-Tagged ABCC4_rs1479390wt: AAAGAAAGAAAGAAAGAAAGTGTA |
| Anti-Tagged ABCC4_rs1479390wt: GTAAAAAGAAAGGTATAAAGGTAA |
| Anti-Tagged ABCC4_rs1151471wt: TTAGTGAAGAAGTATAGTTTATTG |
| Anti-Tagged ABCC4_rs1151471mt: TTGAGTAATTGAATTGTGAAATGA |
| Anti-Tagged ABCC4_rs1059762wt: AAAGAAAGGATTTGTAGTAAGATT |
| Anti-Tagged ABCC4_rs1059762mt: GTAAGTAATGAATGTAAAAGGATT |
| Anti-Tagged ABCC5_ rs1000002wt: ATTGAAAGATGAAAAGATGAAAAG |
| Anti-Tagged ABCC5_ rs1000002mt: GATTGTAAATAGAAAGTGAAGTAA |
| Anti-Tagged ABCC5_ rs4148557wt: GATTTGTAATTGTTGAGTAAATGA |
| Anti-Tagged ABCC5_ rs4148557mt: GATTTGTATTGATTGAGATTAAAG |
| Anti-Tagged ABCC5_ rs3749438wt: AAAGGTAAGATTATTGATGAAAAG |
| Anti-Tagged ABCC5_ rs3749438mt: TGATAAAGTGATAAAGGATTAAAG |
| Anti-Tagged ABCC5_i1-1821wt: TGATTTGAGTATTTGAGATTTTGA |
| Anti-Tagged ABCC5_i1-1821mt: GTATTTAGATGAGTTTGTTAGATT |
| Anti-Tagged ABCC5_i1-1679wt: GTTAGTTAGATTATTGTTAGTTAG |
| Anti-Tagged ABCC5_i1-1679mt: TTAGTGTAGTAAGTTTAAAGTGTA |
| Anti-Tagged ABCC5_i1-1205wt: GTAAGTAAATTGAAAGATTGATGA |
| Anti-Tagged ABCC5_i1-1205mt: AAAGTGAAAAAGATTGATTGATGA |
| Anti-Tagged ABCC5_rs1132776wt: GTTTATAGTGAAATATGAAGATAG |
| Anti-Tagged ABCC5_rs1132776mt: AAAGAAAGATTGTTGAGATTATGA |
| Anti-Tagged ABCC5_rs562wt: GTTAGTTATTGAGAAGTGTATATA |
| Anti-Tagged ABCC5_rs562mt: GTTATGAAATTGTTAGTGTAGATT |
HotStar Taq 2X Master Mix was obtained from Qiagen. Platinum GenoTYPE Tsp DNA polymerase, individual deoxyribonucleoside triphosphates, and biotin-dCTP were purchased from Invitrogen Corporation. Shrimp Alkaline Phosphatase (SAP) was obtained from Promega, Madison, USA and Exonuclease I from New England Biolabs, Beverly, MA, USA. Bioplex coupling beads, sheath fluid, and calibration and validation kits were purchased from Bio-Rad Laboratories, Life Science Research Group, Hercules, CA. The cross-linker 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) was obtained from Pierce, Rockford, IL. The oligonucleotide coupling and hybridization reagents: 2[N-Morpholino] ethanesulfonic acid (MES) and Tris-EDTA buffer were purchased from Sigma Aldrich, NaCl from Merck, 10% sodium dodecyl sulfate (SDS) and Tris from 1st BASE Laboratories Sdn. Bhd, Malaysia, Triton X-100 from BioRad, and Tween 20 from USB, Cleveland, OH. The platinum grade streptavidin-conjugated phycoerythrin (1 g/L) was obtained from Molecular Probes, Eugene, OR, USA.
Whole blood (10 mL) was collected from normal, healthy individuals, aged 18 to 40 years old from a previous study. All volunteer subjects were recruited in accordance with local regulatory and institutional ethics requirements (Institutional Review Board, National University Hospital, Singapore) and written informed consent. Genomic DNA was isolated from whole blood using standard desalting methods. DNA samples were quantified spectrophotometrically by measuring absorbance at 260 nm, diluted to 5 ng/μL, and stored at 4°C.
A total of eight random samples were subjected to DNA sequencing to obtain the genotypic profile of all 22 ABC SNPs in each individual. These include both reported and novel SNPs residing in potentially functional regions of the drug transporters (21-23). For all samples used in the study, genotyping results obtained with the Bioplex assay were compared with those obtained with dideoxy dye-terminator sequencing chemistry. For each of the 22 SNPs, bidirectional (forward and reverse) sequencing was performed using the PCR primers shown in Table 2. DNA sequencing reactions were performed using BigDye Terminator 3.1 Cycle Sequencing Kit and analyzed on the automated ABI Prism Model 3100 Avant Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Multiplex PCR refers to the generation of multiple PCR products in a single reaction. Four separate multiplex PCR panels were required to generate all the fragments containing the 22 SNPs. The primer pairs were designed such that the SNP lies towards the centre of the PCR fragment. The PCR mixtures contained Qiagen Hotstar Taq 1X Master Mix, varying concentrations of primers (Table 2) and 4 ng of genomic DNA in a final volume of 20 μL. Following an initial pre-denaturation step at 95°C for 15 min, the reactions were cycled 35 times through denaturation at 94°C for 1 min, variable annealing temperatures (Table 2) for 1 min and extension at 72°C for 1 min. The reactions were terminated by an additional extension step at 72°C for 10 min. The amplification reactions were performed on the Peltier Thermal Cycler (DNA Engine Dyad; MJ Research Inc, Waltham, MA, USA). The samples were kept at 4°C until use. The PCR products were subjected to 1.6% agarose gel electrophoresis to verify successful amplification of the desired fragments.
Prior to ASPE reaction, 10 μL of each PCR product was treated with 2 U of Exonuclease I and 1 U of SAP by incubating at 37°C for 15 min, followed by enzyme deactivation at 80°C for 20 min. SAP inactivates any remaining nucleotides (particularly dCTP) to allow efficient incorporation of biotin-dCTP during the primer extension reaction and Exonuclease I degrades any remaining PCR primers to avoid interference with the tagged ASPE primers during the extension reaction. The treated samples were then added directly to the ASPE reaction.
Multiplex ASPE reactions comprised 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.25 mM MgCl2, 5 μM biotin-dCTP, 5 μM each of dATP, dGTP, and dTTP, 0.75 U of Platinum GenoTYPE Tsp DNA polymerase, 25 nM each TAG-ASPE primer and 10 μL of pooled treated PCR product in a final volume of 20 μL. The ASPE primer sequences are listed in Table 3. The ASPE reactions were incubated at 96°C for 2 min and then subjected to 35 cycles at 94°C for 30 s, 55°C for 1 min, and 74°C for 2 min. The reactions were then held at 4°C until use.
The aminated anti-tags were coupled to their corresponding populations of carboxylated microspheres using an EDC chemistry (Table 1). Briefly, 2.5 x 106 microspheres of each population were resuspended in 25 μL of 0.1 M MES, pH 4.5, followed by addition of 0.2 nmole of anti-tag oligos. 2.5 μL of freshly-prepared 10 g/L EDC solution was added to each microsphere mixture and incubated for 30 min in the dark. A second 2.5 μL aliquot of freshly prepared EDC solution was added and incubated for an additional 30 min in the dark. Following this, the coupled microspheres were washed in 500 μL of 0.2% Tween 20 and then 500 μL of 0.1% SDS. Finally, the pelleted coupled microspheres were resuspended in 50 μL of TE (10 mM Tris, 1 mM EDTA), pH 8.0. The coupled microspheres were enumerated using a hemacytometer, and stored at 4°C protected from light.
Approximately 2500 microspheres of each of the 44 anti-tag-bearing microsphere sets (Table 1) were combined in 2X Tm Hybridization Buffer [0.4 M NaCl, 0.2 M Tris (pH 8.0), 0.16% Triton X-100]. The microsphere mixture was concentrated and resuspended to 100 of each microsphere set per μL in 2X Tm Hybridization Buffer. A 25 μL aliquot of the mixture was added to each well of a 96-well plate (MJ Research). A 10 μL aliquot of each ASPE reaction and 15 μL sterile water were then added directly to each well. 25 μL of sterile water was added to the background well. The samples were then heated to 96°C for 90 s, followed by 1 hr incubation at 37°C. After this incubation, the samples were washed twice with 1X Tm Hybridization Buffer [0.2 M NaCl, 0.1 M Tris (pH 8.0), 0.08% Triton X-100]. The microspheres were then resuspended in 120 μL of reporter solution (2 mg/L streptavidin-conjugated phycoerythrin in 1X Tm Hybridization Buffer) and incubated for 15 min at 37°C. The reactions were transferred to a flat-bottomed 96-well plate (Nunc) and 50 μL was analyzed on the Luminex xMAP/Bioplex system (Bio-Rad Laboratories, Life Science Research Group, Hercules, CA) at ambient temperature. For each sample, instrument settings were configured to read a minimum of 100 events per bead region with the gate setting being defined before the samples were run and maintained throughout the course of the study.
For each DNA sample subjected to the Bioplex array assay, median fluorescence intensity (MFI) values were generated for each of the 44 microsphere populations corresponding to each allele within the assay. For each allele of a given sample, the NET MFI was set to be the larger of zero and the value obtained by subtracting the no-target (water control) MFI values from the respective MFI values of the sample. The MFI units for at least one allele were required to be at least 10 times the no-target MFI for that allele and magnitude of at least 300. The genotype was then determined based on the mutant allelic ratio where:
EQN01
The mutant allelic ratio is defined as the fraction of the total NET MFI signal for a given SNP attributed to the presence of the mutant allele. Based on pre-set threshold values, the allelic ratio was used to discriminate wild-type, mutant, and heterozygous SNP calls. Threshold values were empirically determined for each individual SNP. The genotype calling was performed with the aid of the Bioplex SNP Manager macro software provided by BioRad [http://www.bio-rad.com/snpmanager].
Results and Discussion
DNA sequencing
A total of eight random DNA samples were sequenced for the genotypic profile of the 22 ABC SNPs as shown in Table 5. Of the 22 SNPs, all three possible genotypes i.e. homozygous wild-type, heterozygous and homozygous mutant were present for 15 of them. Only two genotypes (homozygous wild-type or mutant and heterozygous) were present for six of the SNPs (rs1128503, rs4148557, i1-1679, rs1189437, rs1151471 and rs868853). All the screened subjects were homozygous mutant for one SNP (rs504348) (Table 5). The presence of all three genotypes for the majority (68%) of the SNPs analyzed allows a reliable assessment of the accuracy of the Bioplex suspension array assay.
Table 5.
Genotypic profile of each study subject.
| SNP ID | Subject ID | |||||||
| 01 | 02 | 34 | 35 | 11 | 12 | 13 | 14 | |
| ABCB1_rs1128503 | CT | TT | TT | TT | CT | TT | TT | CT |
| ABCB1_rs2032582 | GT | GT | TT | TT | GT | GG | GT | GA |
| ABCB1_rs1045642 | CT | CT | TT | TT | CT | CT | CC | CC |
| ABCC1_rs504348 | GG | GG | GG | GG | GG | GG | GG | GG |
| ABCC5_ rs1000002 | CC | CT | CT | TT | CC | CC | CC | TT |
| ABCC5_ rs3749438 | GG | GG | AA | GG | GA | AA | AA | GA |
| ABCC5_ rs4148557 | AG | AG | GG | GG | AG | GG | GG | GG |
| ABCC5_rs562 | TT | TC | TC | CC | TT | TT | TT | CC |
| ABCC5_rs1132776 | AA | AG | GG | GG | AG | GG | GG | GG |
| ABCC5_i1-1205 | GA | GG | AA | GA | GA | AA | AA | AA |
| ABCC5_i1-1679 | AT | AA | AA | AT | AA | AA | AA | AT |
| ABCC5_i1-1821 | AA | AA | GG | AA | AA | AA | AA | AG |
| ABCC4_rs1479390 | GT | GT | GG | TT | GG | GG | GG | GG |
| ABCC4_rs2274405 | CC | TC | TT | TC | TT | TC | CC | TC |
| ABCC4_rs1189429 | AA | AA | GA | AA | GA | GG | GA | GA |
| ABCC4_rs4148436 | AG | GG | AG | AA | AG | AG | AG | AG |
| ABCC4_ rs1189437 | TT | TT | TT | TT | GT | TT | TT | TT |
| ABCC4_ rs1151471 | TT | TT | TT | TT | CT | TT | TT | TT |
| ABCC4_ rs1059762 | AA | AA | GA | GA | GG | AA | AA | AA |
| ABCC4_rs868853 | TT | TT | CT | TT | TT | CT | TT | CT |
| ABCC4_rs9524885 | CC | TC | TT | CC | CC | TT | TC | TC |
| ABCC4_rs869951 | GG | CG | CC | GG | GG | CG | CG | CG |
| Wild-type is indicated by the 1st nucleotide of the genotype nomenclature | ||||||||
Multiplex PCR
Multiplex PCR conditions were established in a series of preliminary experiments (data not shown). Multiplex PCRs were optimized for annealing temperatures and individual primer concentrations. A total of 4 multiplex PCR panels were required to generate 21 fragments containing 22 SNPs analyzed in this study (Table 2).
Bioplex suspension array
Tables 6 and 7 show the raw MFI values of each individual sample (assayed in duplicates) and a non-template background control. The results between the duplicates for all readouts were clearly consistent. The genotyping assay for the 22 SNPs was highly specific and robust; the signal-to-noise ratio (specific MFI/background) ranged from >10 to >900 and the MFI units ranged from approximately 300 to 25,000. Threshold values were empirically determined for each individual SNP. The mutant allelic ratio ranged from <0.01-0.28 for wild-type calls, 0.19–0.78 for heterozygous calls, and 0.89–1.00 for mutant calls. While the heterozygotes exhibited almost equal MFI signals for both alleles for the majority of the SNPs, the MFI signal of one allele was higher than the other for a few SNPs. This could possibly be explained by the reasoning that one allele of a SNP amplified disproportionately and obscured the presence of the other allele. Nonetheless, with prior validation using known control samples, the various genotypes are clearly distinguishable. Representative allelic ratio scatterplots are shown for two of the SNPs (Fig. 2).
Fig. 2.
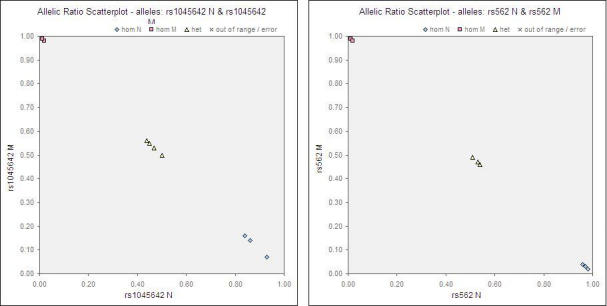
Representative allelic ratio scatterplots (A) rs1045642 and (B) rs562.
Figure 3 depicts the graphical representation of allelic ratio for two of the study subjects. All the 352 genotypes (22 SNPs x 8 subjects x 2 i.e. duplicates) determined by the Bioplex suspension array were 100% concordant with results obtained using the sequencing approach.
Fig. 3.
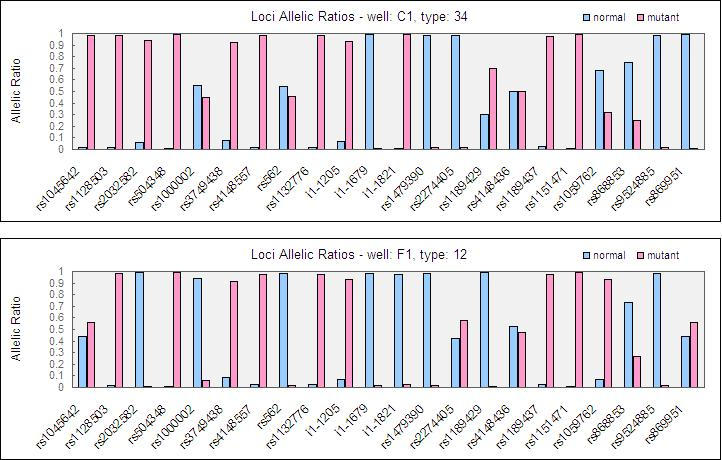
Graphical representation of allelic ratio (A) Subject 34 and (B) Subject 12.
Supplemental Information
This study explores an alternative technological platform, the bead-based suspension array assay to rapidly screen a panel of genetic variants of the ABC transporters, some of which have been previously genotyped for various ethnic groups in our laboratory using minisequencing (21, 22). We have successfully developed and validated a consolidated bead-based genotyping platform for simultaneous detection of 22 SNPs in a single reaction. The results obtained using the Bioplex suspension array were unambiguous and in complete concordance with sequencing. The use of a multiplex PCR approach significantly increased the through-put and cost-savings of the assay. The ASPE method was chosen over single base chain extension and oligonucleotide ligation methods for its ability to read both alleles from a given SNP in a single tube (18, 24, 25). This study is based on a known technology performed on a small sample size. Nonetheless, we believe it is adequate for demonstrating the functionality of the assay as an initial proof-of-concept phase. A more extensive validation study involving large-scale population screening may subsequently be considered for establishing SNP genotyping panels for diagnostic applications.
The Bioplex suspension array offers several advantages over alternative genotyping methodologies. The use of direct sequencing has long been regarded as the “gold standard” for the identification of genetic variations and determination of the nucleotide arrangement in a DNA fragment. However, direct sequencing as a genotyping tool is not a commercially viable option due to its high cost and the laborious procedures involved. Conventional genotyping methods such as allele-specific PCR and restriction fragment length polymorphism have been criticized for their poor sensitivity and specificity, are labour-intensive and time consuming. The sophisticated Affymetrix Genechip microarray, though powerful and high-throughput, is very expensive and serves more as a fishing expedition than a simple genotyping exercise catered for a specific application. The microarray is an investigative approach used widely for the search of candidate genes associated with a disease state. The Bioplex platform, on the other hand, serves as a diagnostic tool to complement microarray analysis by targeting the set of genes identified from the search. A predefined SNP genotyping assay for specific applications ensures a standardized format for rapid and cost-effective genetic analysis.
The Bioplex platform allows for the simultaneous detection of up to 50 SNPs in a single reaction well. At present, we have proven it to be a robust, reliable and reasonably high-throughput genotyping tool for multiplex analysis of 22 SNPs. The current system interface allows analysis of 2 x 96 = 192 samples in a single day, equivalent to a maximum output of 9,600 genotype designations. This technology positions itself strategically between the conventional genotyping techniques and the high-end oligonucleotide arrays. Most importantly, its versatility enables the addition of new SNPs or removal of unwanted SNPs to create customized genotyping panels for the consumer. The Bioplex platform allows for all genotyping reactions to be completed and maintained in a single multiplex assay which enables better quality assurance to be achieved. Some other appealing factors of the Bioplex platform include its requirement for minimal DNA template, true liquid phase kinetics, short running time, ease of data analysis and affordability. The disadvantages of the platform include its ability to only detect known mutations, limited throughput due to the current commercially available beads, the need for empirical determination of the threshold value for genotype designation, and the high start-up cost.
This technology offers a promising genotyping platform for developing validated multiplex genotyping kits for specific commercial and research applications involving large-scale mutation screening. There is rising awareness among pharmaceutical companies and clinicians on the importance of pharmacogenetics. The growing trend of pharmaceutical companies integrating the pharmacogenetics programme in clinical trials during new drug development is becoming evident. In view of the emerging field of pharmacogenetics, it is anticipated that there will be a high demand for rapid, reliable and economical genotyping services from pharmaceutical companies, research institutions, hospitals and clinics.
The protocol that we are presenting will indeed be helpful to researchers in the field or to newcomers. This report provides the technical information necessary for establishing SNP panel testing using the Bioplex suspension array. Some relevant areas for this application include genes that impact on drug disposition/therapeutic response or disease susceptibility. These SNP panels can be classified as a combination of markers that affect response to a given drug e.g. a chemotherapeutic agent, those that confer a risk for pathologies for which prophylactic therapies are available, or those that can be used to complement clinical diagnosis of diseases. The application of this strategy in therapeutics and disease association studies will eventually lead to routine adoption of the platform in clinical molecular diagnostics.
TABLES
Table 6.
MFI values of each microsphere set corresponding to each allele of ABCB1, ABCC1 and ABCC5.
N: normal, M: mutant, blue: homozygous wild-type, yellow: heterozygous, pink: homozygous mutant The SNPs are denoted by the last 3-4 digits of their respective full ID.
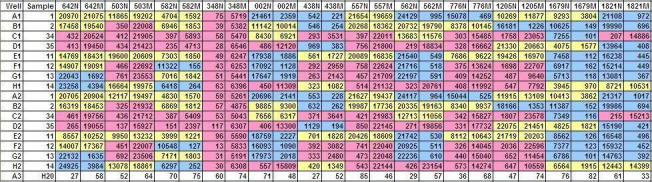 |
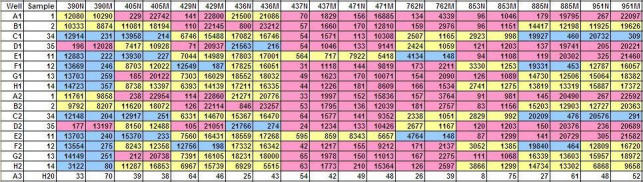
Table 7.
MFI values of each microsphere set corresponding to each allele of ABCC4.
N: normal, M: mutant, blue: homozygous wild-type, yellow: heterozygous, pink: homozygous mutant The SNPs are denoted by the last 3 digits of their respective full ID.
 |
Acknowledgments
This work was funded by the cross-faculty research grant, “Niche Area Programme,” National University of Singapore, Singapore.
Abbreviations
- ABC
ATP-binding cassette
- ASPE
allele-specific primer extension
- EDC
1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride
- MES
2[N-Morpholino] ethanesulfonic acid
- MFI
median fluorescence intensity
- SAP
shrimp alkaline phosphatase
- SDS
sodium dodecyl sulfate
- SNP
single nucleotide polymorphism
Protocols
Microspheres should be protected from prolonged exposure to light throughout the following procedures.
Microsphere coupling
Bring a fresh aliquot of -20°C, desiccated EDC powder to room temperature.
Resuspend the amine-substituted oligonucleotide (“probe” or “capture” oligo) to 100 μM in sterile water.
Resuspend the stock microspheres by vortexing for approximately 20 seconds.
Transfer 2.5 x 106 of the stock microspheres to a microfuge tube.
Pellet the stock microspheres by microcentrifugation at 10,000 x g for 3 minutes.
Remove the supernatant and resuspend the pelleted microspheres in 25 μL of 0.1 M MES, pH 4.5 by vortexing for approximately 20 seconds.
Add 2 μL of the capture oligo (100 μM) to the resuspended microspheres and mix by vortexing.
Prepare a fresh solution of 10 g/L EDC in sterile water. (Note: Return the EDC powder to desiccant to re-use for the second EDC addition.)
One by one for each reaction, add 2.5 μL of fresh 10 g/L EDC to the microspheres and mix by vortexing.
Incubate for 30 minutes at room temperature in the dark.
Prepare a second fresh solution of 10 g/L EDC in sterile water.
One by one for each reaction, add 2.5 μL of fresh 10 g/L EDC to the microspheres and mix by vortexing.
Incubate for 30 minutes at room temperature in the dark.
Add 500 μL of 0.02% Tween-20 to the coupled microspheres.
Pellet the coupled microspheres by microcentrifugation at 10,000 x g for 3 minutes.
Remove the supernatant and resuspend the coupled microspheres in 500 μL of 0.1% SDS by vortexing.
Pellet the coupled microspheres by microcentrifugation at 10,000 x g for 3 minutes.
Remove the supernatant and resuspend the coupled microspheres in 50 μL of TE, pH 8.0 by vortexing for approximately 20 seconds.
-
Enumerate the coupled microspheres by hemacytometer:
- Dilute the resuspended, coupled microspheres 1:100 in sterile water.
- Mix thoroughly by vortexing.
- Transfer 10 μL to the hemacytometer.
- Count the microspheres within the 4 large corners of the hemacytometer grid.
- Microspheres/μL = (Sum of microspheres in 4 large corners) x 2.5 x 100 (dilution factor). (Note: maximum is 50,000 microspheres/μL.)
Store coupled microspheres refrigerated at 4°C in the dark.
Hybridisation and detection
Select the appropriate microsphere sets and resuspend by vortexing for approximately 20 seconds.
Combine 2500 microspheres of each set per reaction.
Concentrate the microsphere mixture by centrifugation at 10,000 x g for 3 minutes.
Remove the supernatant and resuspend to 100 of each microsphere set per μL in 2X Tm Hybridization Buffer by vortexing for approximately 20 seconds.
Aliquot 25 μL of the microsphere mixture to each well.
Add 25 μL of sterile water to each background well.
Add 10 μL of each ASPE reaction to appropriate wells. (Note: 5 μL is usually sufficient.)
Adjust the total volume to 50 μL by adding 15 μL of sterile water to each sample well.
Cover the plate to prevent evaporation and denature at 96°C for 90 seconds.
Hybridize at 37°C for 1 hour.
Pellet the microspheres by centrifugation at 5000 x g for 3 minutes and remove the supernatant.
Resuspend the pelleted microspheres in 70 μL of 1X Tm Hybridization Buffer.
Pellet the microspheres by centrifugation at 5000 x g for 3 minutes and remove the supernatant.
Repeat steps 11 and 12 for a total of two washes.
Resuspend microspheres in 120 μL of 1X Tm Hybridization Buffer containing 2 mg/L streptavidin-R-phycoerythrin.
Incubate at 37°C for 15 minutes.
Analyze 50 °L at room temperature on the Luminex xMAP/Bioplex analyzer.
References
- Pharmacogenomics: the inherited basis for interindividual differences in drug response. Evans WE, Johnson JA. Annu Rev Genomics Hum Genet. 2001;2:9–39. doi: 10.1146/annurev.genom.2.1.9. [DOI] [PubMed] [Google Scholar]
- Pharmacogenomics: translating functional genomics into rational therapeutics. Evans WE, Relling MV. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- Genetic variation as a guide to drug development. Kleyn PW, Vesell ES. Science. 1998;281:1820–1821. doi: 10.1126/science.281.5384.1820. [DOI] [PubMed] [Google Scholar]
- Pharmacogenomics: state of the research and perspectives in clinical application. Severino G, Chillotti C, Stochino ME, Del Zompo M. Neurol Sci. 2003;24:S146–148. doi: 10.1007/s100720300064. [DOI] [PubMed] [Google Scholar]
- Pharmacogenetics approach to therapeutics. Koo SH, Lee EJ. Clin Exp Pharmacol Physiol. 2006;33:525–532. doi: 10.1111/j.1440-1681.2006.04402.x. [DOI] [PubMed] [Google Scholar]
- Four-digit allele genotyping of the HLA-A and HLA-B genes in Japanese patients with Behcet's disease by a PCR-SSOP-Luminex method. Itoh Y, Inoko H, Kulski JK, Sasaki S, Meguro A, Takiyama N, Nishida T, Yuasa T, Ohno S, Mizuki N. Tissue Antigens. 2006;67:360–364. doi: 10.1111/j.1399-0039.2006.00586.x. [DOI] [PubMed] [Google Scholar]
- High-throughput DNA typing of HLA-A, -B, -C, and -DRB1 loci by a PCR-SSOP-Luminex method in the Japanese population. Itoh Y, Mizuki N, Shimada T, Azuma F, Itakura M, Kashiwase K, Kikkawa E, Kulski JK, Satake M, Inoko H. Immunogenetics. 2005;57:717–729. doi: 10.1007/s00251-005-0048-3. [DOI] [PubMed] [Google Scholar]
- Multiplex genotyping of human minor histocompatibility antigens. Pietz BC, Warden MB, DuChateau BK, Ellis TM. Hum Immunol. 2005;66:1174–1182. doi: 10.1016/j.humimm.2005.08.243. [DOI] [PubMed] [Google Scholar]
- Flow cytometric assay for genotyping cytochrome p450 2C9 and 2C19: comparison with a microelectronic DNA array. Pickering JW, McMillin GA, Gedge F, Hill HR, Lyon E. Am J Pharmacogenomics. 2004;4:199–207. doi: 10.2165/00129785-200404030-00007. [DOI] [PubMed] [Google Scholar]
- Simultaneous determination of 7 N-acetyltransferase-2 single-nucleotide variations by allele-specific primer extension assay. Zhu Y, Hein DW, Doll MA, Reynolds KK, Abudu N, Valdes Jr. R, Linder MW. Clin Chem. 2006;52:1033–1039. doi: 10.1373/clinchem.2005.063198. [DOI] [PubMed] [Google Scholar]
- Analytical validation of the tag-it high-throughput microsphere-based universal array genotyping platform: application to the multiplex detection of a panel of thrombophilia-associated single-nucleotide polymorphisms. Bortolin S, Black M, Modi H, Boszko I, Kobler D, Fieldhouse D, Lopes E, Lacroix JM, Grimwood R, Wells P, Janeczko R, Zastawny R. Clin Chem. 2004;50:2028–2036. doi: 10.1373/clinchem.2004.035071. [DOI] [PubMed] [Google Scholar]
- Multiplexed genotyping of beta-globin variants from PCR-amplified newborn blood spot DNA by hybridization with allele-specific oligodeoxynucleotides coupled to an array of fluorescent microspheres. Colinas RJ, Bellisario R, Pass KA. Clin Chem. 2000;46:996–998. [PubMed] [Google Scholar]
- High-throughput detection of pathogenic yeasts of the genus trichosporon. Diaz MR, Fell JW. J Clin Microbiol. 2004;42:3696–3706. doi: 10.1128/JCM.42.8.3696-3706.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technical validation of a multiplex platform to detect thirty mutations in eight genetic diseases prevalent in individuals of Ashkenazi Jewish descent. Strom CM, Janeczko RA, Anderson B, Redman J, Quan F, Buller A, McGinniss MJ, Sun WM. Genet Med. 2005;7:63–639. doi: 10.1097/01.gim.0000187120.93597.16. [DOI] [PubMed] [Google Scholar]
- Transfer of a Mycobacterium tuberculosis genotyping method, Spoligotyping, from a reverse line-blot hybridization, membrane-based assay to the Luminex multianalyte profiling system. Cowan LS, Diem L, Brake MC, Crawford JT. J Clin Microbiol. 2004;42:474–477. doi: 10.1128/JCM.42.1.474-477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genotyping of human papillomavirus in cervical lesions by L1 consensus PCR and the Luminex xMAP system. Jiang HL, Zhu HH, Zhou LF, Chen F, Chen Z. J Med Microbiol. 2006;55:715–720. doi: 10.1099/jmm.0.46493-0. [DOI] [PubMed] [Google Scholar]
- Bead-based multiplex genotyping of human papillomaviruses. Schmitt M, Bravo IG, Snijders PJ, Gissmann L, Pawlita M, Waterboer T. J Clin Microbiol. 2006;44:504–512. doi: 10.1128/JCM.44.2.504-512.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluorescent microsphere-based readout technology for multiplexed human single nucleotide polymorphism analysis and bacterial identification. Ye F, Li MS, Taylor JD, Nguyen Q, Colton HM, Casey WM, Wagner M, Weiner MP, Chen J. Hum Mutat. 2001;17:305–316. doi: 10.1002/humu.28. [DOI] [PubMed] [Google Scholar]
- Quantitative, multiplexed detection of bacterial pathogens: DNA and protein applications of the Luminex LabMAP system. Dunbar SA, Vander Zee CA, Oliver KG, Karem KL, Jacobson JW. J Microbiol Methods. 2003;53:245–252. doi: 10.1016/S0167-7012(03)00028-9. [DOI] [PubMed] [Google Scholar]
- A bead-based method for multiplexed identification and quantitation of DNA sequences using flow cytometry. Spiro A, Lowe M, Brown D. Appl Environ Microbiol. 2000;66:4258–4265. doi: 10.1128/AEM.66.10.4258-4265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simultaneous genotyping of seven single-nucleotide polymorphisms in the MDR1 gene by single-tube multiplex minisequencing. Gwee PC, Tang K, Chua JM, Lee EJ, Chong SS, Lee CG. Clin Chem. 2003;49:672–676. doi: 10.1373/49.4.672. [DOI] [PubMed] [Google Scholar]
- Strong linkage disequilibrium at the nucleotide analogue transporter ABCC5 gene locus. Gwee PC, Tang K, Sew PH, Lee EJ, Chong SS, Lee CG. Pharmacogenet Genomics. 2005;15:91–104. doi: 10.1097/01213011-200502000-00005. [DOI] [PubMed] [Google Scholar]
- A functional polymorphism within the MRP1 gene locus identified through its genomic signature of positive selection. Wang Z, Wang B, Tang K, Lee EJ, Chong SS, Lee CG. Hum Mol Genet. 2005;14:2075–2087. doi: 10.1093/hmg/ddi212. [DOI] [PubMed] [Google Scholar]
- Comparison of four flow cytometric SNP detection assays and their use in plant improvement. Lee SH, Walker DR, Cregan PB, Boerma HR. Theor Appl Genet. 2004;110:167–174. doi: 10.1007/s00122-004-1827-1. [DOI] [PubMed] [Google Scholar]
- Flow cytometric platform for high-throughput single nucleotide polymorphism analysis. Taylor JD, Briley D, Nguyen Q, Long K, Iannone MA, Li MS, Ye F, Afshari A, Lai E, Wagner M, Chen J, Weiner MP. Biotechniques. 2001;30:661–669. doi: 10.2144/01303dd04. [DOI] [PubMed] [Google Scholar]


